Structure, mechanism, and inhibition of the zinc-dependent histone deacetylases
- PMID: 30743180
- PMCID: PMC6687579
- DOI: 10.1016/j.sbi.2019.01.004
Structure, mechanism, and inhibition of the zinc-dependent histone deacetylases
Abstract
Zinc-dependent histone deacetylases (HDACs) regulate the biological function of histone and non-histone proteins through the hydrolysis of acetyllysine side chains to yield free lysine and acetate. Certain HDAC isozymes exhibit alternative catalytic activities, such as polyamine deacetylase or lysine fatty acid deacylase activity. To date, crystal structures have been reported for class I HDACs (1, 2, 3, and 8), class IIa HDACs (4 and 7), and class IIb HDACs (6 and 10). Conserved active site residues mediate the chemistry of substrate activation and hydrolysis in these isozymes through a metal-activated water molecule assisted by general base-general acid catalysis. Upregulated HDAC activity is observed in cancer and neurodegenerative disease, and four HDAC inhibitors are currently approved for use in cancer chemotherapy. Crystal structures of HDAC-inhibitor complexes guide the design of new inhibitors with high affinity and selectivity for specific HDAC isozymes implicated in human disease.
Conflict of interest statement
Conflict of interest
Nothing declared.
Figures


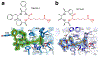
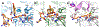

References
-
- Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M: The growing landscape of lysine acetylation links metabolism and cell signaling. Nat Rev Mol Cell Biol 2014, 15:536–550. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

