Metabolic Reprogramming of the Host Cell by Human Adenovirus Infection
- PMID: 30744016
- PMCID: PMC6409786
- DOI: 10.3390/v11020141
Metabolic Reprogramming of the Host Cell by Human Adenovirus Infection
Abstract
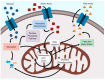
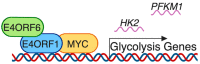
Viruses are obligate intracellular parasites that alter many cellular processes to create an environment optimal for viral replication. Reprogramming of cellular metabolism is an important, yet underappreciated feature of many viral infections, as this ensures that the energy and substrates required for viral replication are available in abundance. Human adenovirus (HAdV), which is the focus of this review, is a small DNA tumor virus that reprograms cellular metabolism in a variety of ways. It is well known that HAdV infection increases glucose uptake and fermentation to lactate in a manner resembling the Warburg effect observed in many cancer cells. However, HAdV infection induces many other metabolic changes. In this review, we integrate the findings from a variety of proteomic and transcriptomic studies to understand the subtleties of metabolite and metabolic pathway control during HAdV infection. We review how the E4ORF1 protein of HAdV enacts some of these changes and summarize evidence for reprogramming of cellular metabolism by the viral E1A protein. Therapies targeting altered metabolism are emerging as cancer treatments, and similar targeting of aberrant components of virally reprogrammed metabolism could have clinical antiviral applications.
Keywords: E1A; E4ORF1; HAdV36; HAdV5; MYC; Warburg effect; glutaminolysis; glycolysis; human adenovirus; metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Nash L.A., Parks R.J. Adenovirus Biology and Development as a Gene Delivery Vector. In: Ng P., Brunetti-Pierri N., editors. Therapeutic Applications of Adenoviruses. CRC Press; Boca Raton, FL, USA: 2017. pp. 1–36.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

