BAG3 and SYNPO (synaptopodin) facilitate phospho-MAPT/Tau degradation via autophagy in neuronal processes
- PMID: 30744518
- PMCID: PMC6613891
- DOI: 10.1080/15548627.2019.1580096
BAG3 and SYNPO (synaptopodin) facilitate phospho-MAPT/Tau degradation via autophagy in neuronal processes
Abstract
A major cellular catabolic pathway in neurons is macroautophagy/autophagy, through which misfolded or aggregation-prone proteins are sequestered into autophagosomes that fuse with lysosomes, and are degraded. MAPT (microtubule-associated protein tau) is one of the protein clients of autophagy. Given that accumulation of hyperphosphorylated MAPT contributes to the pathogenesis of Alzheimer disease and other tauopathies, decreasing endogenous MAPT levels has been shown to be beneficial to neuronal health in models of these diseases. A previous study demonstrated that the HSPA/HSP70 co-chaperone BAG3 (BCL2-associated athanogene 3) facilitates endogenous MAPT clearance through autophagy. These findings prompted us to further investigate the mechanisms underlying BAG3-mediated autophagy in the degradation of endogenous MAPT. Here we demonstrate for the first time that BAG3 plays an important role in autophagic flux in the neurites of mature neurons (20-24 days in vitro [DIV]) through interaction with the post-synaptic cytoskeleton protein SYNPO (synaptopodin). Loss of either BAG3 or SYNPO impeded the fusion of autophagosomes and lysosomes predominantly in the post-synaptic compartment. A block of autophagy leads to accumulation of the autophagic receptor protein SQSTM1/p62 (sequestosome 1) as well as MAPT phosphorylated at Ser262 (p-Ser262). Furthermore, p-Ser262 appears to accumulate in autophagosomes at post-synaptic densities. Overall these data provide evidence of a novel role for the co-chaperone BAG3 in synapses. In cooperation with SYNPO, it functions as part of a surveillance complex that facilitates the autophagic clearance of MAPT p-Ser262, and possibly other MAPT species at the post-synapse. This appears to be crucial for the maintenance of a healthy, functional synapse.Abbreviations: aa: amino acids; ACTB: actin beta; BafA1: bafilomycin A1; BAG3: BCL2 associated athanogene 3; CQ chloroquine; CTSL: cathepsin L; DIV: days in vitro; DLG4/PSD95: discs large MAGUK scaffold protein 4; HSPA/HSP70: heat shock protein family A (Hsp70); MAP1LC3B/LC3B: microtubule associated protein 1 light chain 3 beta; MAP2: microtubule associated protein 2; MAPT: microtubule associated protein tau; p-Ser262: MAPT phosphorylated at serine 262; p-Ser396/404: MAPT phosphorylated at serines 396 and 404; p-Thr231: MAPT phosphorylated at threonine 231; PBS: phosphate buffered saline; PK: proteinase K; scr: scrambled; shRNA: short hairpin RNA; SQSTM1/p62 sequestosome 1; SYN1: synapsin I; SYNPO synaptopodin; SYNPO2/myopodin: synaptopodin 2; VPS: vacuolar protein sorting.
Keywords: Autophagosome; PPxY domain; SQSTM1/p62; WW domain; postsynaptic density; synapse.
Figures







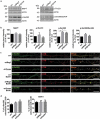
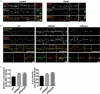
Similar articles
-
MAPT/Tau accumulation represses autophagy flux by disrupting IST1-regulated ESCRT-III complex formation: a vicious cycle in Alzheimer neurodegeneration.Autophagy. 2020 Apr;16(4):641-658. doi: 10.1080/15548627.2019.1633862. Epub 2019 Jun 28. Autophagy. 2020. PMID: 31223056 Free PMC article.
-
Decrease of neuronal FKBP4/FKBP52 modulates perinuclear lysosomal positioning and MAPT/Tau behavior during MAPT/Tau-induced proteotoxic stress.Autophagy. 2021 Nov;17(11):3491-3510. doi: 10.1080/15548627.2021.1875611. Epub 2021 Jan 25. Autophagy. 2021. PMID: 33459145 Free PMC article.
-
The cargo receptor SQSTM1 ameliorates neurofibrillary tangle pathology and spreading through selective targeting of pathological MAPT (microtubule associated protein tau).Autophagy. 2019 Apr;15(4):583-598. doi: 10.1080/15548627.2018.1532258. Epub 2018 Oct 16. Autophagy. 2019. PMID: 30290707 Free PMC article.
-
Commentary: BAG3 as a Mediator of Endosome Function and Tau Clearance.Neuroscience. 2023 May 10;518:4-9. doi: 10.1016/j.neuroscience.2022.05.002. Epub 2022 May 10. Neuroscience. 2023. PMID: 35550160 Free PMC article. Review.
-
Regulation of selective autophagy: the p62/SQSTM1 paradigm.Essays Biochem. 2017 Dec 12;61(6):609-624. doi: 10.1042/EBC20170035. Print 2017 Dec 12. Essays Biochem. 2017. PMID: 29233872 Review.
Cited by
-
Tau Post-translational Modifications: Dynamic Transformers of Tau Function, Degradation, and Aggregation.Front Neurol. 2021 Jan 7;11:595532. doi: 10.3389/fneur.2020.595532. eCollection 2020. Front Neurol. 2021. PMID: 33488497 Free PMC article. Review.
-
The NDR family of kinases: essential regulators of aging.Front Mol Neurosci. 2024 May 13;17:1371086. doi: 10.3389/fnmol.2024.1371086. eCollection 2024. Front Mol Neurosci. 2024. PMID: 38803357 Free PMC article. Review.
-
The role of BAG3 in health and disease: A "Magic BAG of Tricks".J Cell Biochem. 2022 Jan;123(1):4-21. doi: 10.1002/jcb.29952. Epub 2021 May 14. J Cell Biochem. 2022. PMID: 33987872 Free PMC article. Review.
-
Multi-omics analysis identifies RFX7 targets involved in tumor suppression and neuronal processes.Cell Death Discov. 2023 Mar 3;9(1):80. doi: 10.1038/s41420-023-01378-1. Cell Death Discov. 2023. PMID: 36864036 Free PMC article.
-
Autophagic Pathways to Clear the Tau Aggregates in Alzheimer's Disease.Cell Mol Neurobiol. 2021 Aug;41(6):1175-1181. doi: 10.1007/s10571-020-00897-0. Epub 2020 Jun 11. Cell Mol Neurobiol. 2021. PMID: 32529542 Free PMC article. Review.
References
-
- Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. - PubMed
-
- Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
