Comparative transcriptome analysis reveals K+ transporter gene contributing to salt tolerance in eggplant
- PMID: 30744551
- PMCID: PMC6371450
- DOI: 10.1186/s12870-019-1663-8
Comparative transcriptome analysis reveals K+ transporter gene contributing to salt tolerance in eggplant
Abstract
Background: Soil salinization is one of the most crucial abiotic stresses that limit the growth and production of eggplant. The existing researches in eggplant were mostly focused on salt-induced morphological, biochemical and physiological changes, with only limited works centered on salt-response genes in eggplant at the transcriptomic level.
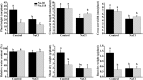
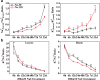
Results: Our preliminary work found that Zhusiqie (No.118) is salt-tolerant and Hongqie (No.30) is salt-sensitive. Consequently, they were re-named as ST118 and SS30, respectively. ST118 showed less damaged on growth and higher K+/Na+ ratios in leaves than SS30. Comparative-transcriptome analysis was used as a powerful approach to understand the salt-response mechanisms in the leaves and roots of SS30 and ST118. And it revealed that genotype-specific and organ-specific manners exist in eggplant in response to salt stress. Strikingly, the genotype-specific differentially expressed genes (DEGs) in ST118 were considered crucial to its higher salt-tolerance, because the expression patterns of common DEGs in the leaves/roots of the two eggplant genotypes were almost the same. Among them, some transcription factors have been reported to be in response to elevated external salinity, including the members of C2C2-CO-like, WRKY, MYB and NAC family. In addition, the AKT1, KAT1 and SOS1 were up-regulated only in the leaves of ST118. Furthermore, the complementation assays demonstrated that the salt-tolerances of both yeast and Arabidopsis akt1 mutants were enhanced by heterologous expression of SmAKT1.
Conclusion: The comparative-transcriptome analysis indicated that the salt-tolerance can be increased by higher transcript level of some genotype-specific genes. This work revealed that eggplants seem to be more inclined to absorb K+ rather than to exclude Na+ under salt stress conditions because seven K+ transporters were significantly up-regulated, while only one Na+ transporter was similarly regulated. Finally, the complementation assays of SmAKT1, which is genotype-specific up-regulated in ST118, suggest that the other TFs and K+ transport genes were worthy of future investigation for their functions in salinity tolerance.
Keywords: Comparative-transcriptome; Eggplant (Solanum melongena L.); Genotype-specific expression; Salt stress; SmAKT1.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable, as this study did not involve human or animal subjects, and the seeds of two eggplant cultivars were stored in School of Agriculture and Biology, Shanghai Jiao Tong University. The seeds of Arabidopsis
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




Similar articles
-
Transcriptome profiling reveals key genes in eggplant (Solanum melongena) roots under salt stress.BMC Genomics. 2025 Jul 4;26(1):635. doi: 10.1186/s12864-025-11802-8. BMC Genomics. 2025. PMID: 40615784 Free PMC article.
-
Comparative analysis of alfalfa (Medicago sativa L.) leaf transcriptomes reveals genotype-specific salt tolerance mechanisms.BMC Plant Biol. 2018 Feb 15;18(1):35. doi: 10.1186/s12870-018-1250-4. BMC Plant Biol. 2018. PMID: 29448940 Free PMC article.
-
Linking genetic determinants with salinity tolerance and ion relationships in eggplant, tomato and pepper.Sci Rep. 2021 Aug 11;11(1):16298. doi: 10.1038/s41598-021-95506-5. Sci Rep. 2021. PMID: 34381090 Free PMC article.
-
Identification and characterization of CBL and CIPK gene families in eggplant (Solanum melongena L.).Mol Genet Genomics. 2016 Aug;291(4):1769-81. doi: 10.1007/s00438-016-1218-8. Epub 2016 Jun 10. Mol Genet Genomics. 2016. PMID: 27287616
-
Salinity-induced expression of HKT may be crucial for Na(+) exclusion in the leaf blade of huckleberry (Solanum scabrum Mill.), but not of eggplant (Solanum melongena L.).Biochem Biophys Res Commun. 2015 May 1;460(2):416-21. doi: 10.1016/j.bbrc.2015.03.048. Epub 2015 Mar 18. Biochem Biophys Res Commun. 2015. PMID: 25796329
Cited by
-
Analysis of Gene Expression Changes in Plants Grown in Salty Soil in Response to Inoculation with Halophilic Bacteria.Int J Mol Sci. 2021 Mar 31;22(7):3611. doi: 10.3390/ijms22073611. Int J Mol Sci. 2021. PMID: 33807153 Free PMC article. Review.
-
Identification and Characterization of Shaker Potassium Channel Gene Family and Response to Salt and Chilling Stress in Rice.Int J Mol Sci. 2024 Sep 8;25(17):9728. doi: 10.3390/ijms25179728. Int J Mol Sci. 2024. PMID: 39273675 Free PMC article.
-
Integrated transcriptome and miRNA sequencing approaches provide insights into salt tolerance in allotriploid Populus cathayana.Planta. 2021 Jul 5;254(2):25. doi: 10.1007/s00425-021-03600-9. Planta. 2021. PMID: 34226949
-
Transcriptome profiling reveals key genes in eggplant (Solanum melongena) roots under salt stress.BMC Genomics. 2025 Jul 4;26(1):635. doi: 10.1186/s12864-025-11802-8. BMC Genomics. 2025. PMID: 40615784 Free PMC article.
-
Impact of Grafting, Salinity and Irrigation Water Composition on Eggplant Fruit Yield and Ion Relations.Sci Rep. 2019 Dec 18;9(1):19373. doi: 10.1038/s41598-019-55841-0. Sci Rep. 2019. PMID: 31853094 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

