Generation of Pigs Resistant to Highly Pathogenic-Porcine Reproductive and Respiratory Syndrome Virus through Gene Editing of CD163
- PMID: 30745836
- PMCID: PMC6367541
- DOI: 10.7150/ijbs.25862
Generation of Pigs Resistant to Highly Pathogenic-Porcine Reproductive and Respiratory Syndrome Virus through Gene Editing of CD163
Abstract
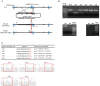
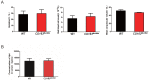
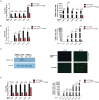
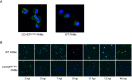
Porcine reproductive and respiratory syndrome (PRRS) is a highly contagious disease and the most economically important disease of the swine industry worldwide. Highly pathogenic-PRRS virus (HP-PRRSV) is a variant of PRRSV, which caused high morbidity and mortality. Scavenger receptor CD163, which contains nine scavenger receptor cysteine-rich (SRCR) domains, is a key entry mediator for PRRSV. A previous study demonstrated that SRCR domain 5 (SRCR5), encoded by exon 7, was essential for PRRSV infection in vitro. Here, we substituted exon 7 of porcine CD163 with the corresponding exon of human CD163-like 1 (hCD163L1) using a CRISPR/Cas9 system combined with a donor vector. In CD163Mut/Mut pigs, modifying CD163 gene had no adverse effects on hemoglobin-haptoglobin (Hb-Hp) complex clearance or erythroblast growth. In vitro infection experiments showed that the CD163 mutant strongly inhibited HP-PRRSV replication by inhibiting virus uncoating and genome release. Compared to wild-type (WT) pigs in vivo, HP-PRRSV-infected CD163Mut/Mut pigs showed a substantially decreased viral load in blood and relief from PRRSV-induced fever. While all WT pigs were dead, there of four CD163Mut/Mut pigs survived and recovered at the termination of the experiment. Our data demonstrated that modifying CD163 remarkably inhibited PRRSV replication and protected pigs from HP-PRRSV infection, thus establishing a good foundation for breeding PRRSV-resistant pigs via gene editing technology.
Keywords: CD163; CRISPR/Cas9; HP-PRRSV.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

References
-
- Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH. et al. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. Journal of the American Veterinary Medical Association. 2005;227(3):385–92. - PubMed
-
- Nieuwenhuis N, Duinhof TF, van Nes A. Economic analysis of outbreaks of porcine reproductive and respiratory syndrome virus in nine sow herds. The Veterinary record. 2012;170(9):225. - PubMed
-
- Rossow KD. Porcine reproductive and respiratory syndrome. Veterinary pathology. 1998;35(1):1–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

