Dalmanol biosyntheses require coupling of two separate polyketide gene clusters
- PMID: 30746075
- PMCID: PMC6335865
- DOI: 10.1039/c8sc03697g
Dalmanol biosyntheses require coupling of two separate polyketide gene clusters
Abstract
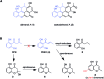
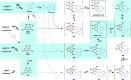
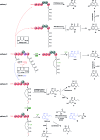
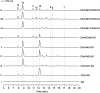
Polyketide-polyketide hybrids are unique natural products with promising bioactivity, but the hybridization processes remain poorly understood. Herein, we present that the biosynthetic pathways of two immunosuppressants, dalmanol A and acetodalmanol A, result from an unspecific monooxygenase triggered hybridization of two distinct polyketide (naphthalene and chromane) biosynthetic gene clusters. The orchestration of the functional dimorphism of the polyketide synthase (ChrA) ketoreductase (KR) domain (shortened as ChrA KR) with that of the KR partner (ChrB) in the bioassembly line increases the polyketide diversity and allows the fungal generation of plant chromanes (e.g., noreugenin) and phloroglucinols (e.g., 2,4,6-trihydroxyacetophenone). The simultaneous fungal biosynthesis of 1,3,6,8- and 2-acetyl-1,3,6,8-tetrahydroxynaphthalenes was addressed as well. Collectively, the work may symbolize a movement in understanding the multiple-gene-cluster involved natural product biosynthesis, and highlights the possible fungal generations of some chromane- and phloroglucinol-based phytochemicals.
Figures

Similar articles
-
Biosynthesis of Fungal Natural Products Involving Two Separate Pathway Crosstalk.J Fungi (Basel). 2022 Mar 21;8(3):320. doi: 10.3390/jof8030320. J Fungi (Basel). 2022. PMID: 35330322 Free PMC article. Review.
-
Natural biocombinatorics in the polyketide synthase genes of the actinobacterium Streptomyces avermitilis.PLoS Comput Biol. 2006 Oct 6;2(10):e132. doi: 10.1371/journal.pcbi.0020132. Epub 2006 Aug 21. PLoS Comput Biol. 2006. PMID: 17029557 Free PMC article.
-
[Progress in fungal polyketide biosynthesis].Sheng Wu Gong Cheng Xue Bao. 2018 Feb 25;34(2):151-164. doi: 10.13345/j.cjb.170219. Sheng Wu Gong Cheng Xue Bao. 2018. PMID: 29424130 Review. Chinese.
-
Sequence-based classification of type II polyketide synthase biosynthetic gene clusters for antiSMASH.J Ind Microbiol Biotechnol. 2019 Mar;46(3-4):469-475. doi: 10.1007/s10295-018-02131-9. Epub 2019 Jan 4. J Ind Microbiol Biotechnol. 2019. PMID: 30610412
-
Expanding our understanding of sequence-function relationships of type II polyketide biosynthetic gene clusters: bioinformatics-guided identification of Frankiamicin A from Frankia sp. EAN1pec.PLoS One. 2015 Apr 2;10(4):e0121505. doi: 10.1371/journal.pone.0121505. eCollection 2015. PLoS One. 2015. PMID: 25837682 Free PMC article.
Cited by
-
Galewone, an Anti-Fibrotic Polyketide from Daldinia eschscholzii with an Undescribed Carbon Skeleton.Sci Rep. 2019 Oct 4;9(1):14316. doi: 10.1038/s41598-019-50868-9. Sci Rep. 2019. PMID: 31586120 Free PMC article.
-
Natural Occurrence of Hybrid Polyketides from Two Distinct Biosynthetic Pathways in Streptomyces pactum.ACS Chem Biol. 2021 Feb 19;16(2):270-276. doi: 10.1021/acschembio.0c00982. Epub 2021 Feb 8. ACS Chem Biol. 2021. PMID: 33601889 Free PMC article.
-
Biosynthesis of 6-Hydroxymellein Requires a Collaborating Polyketide Synthase-like Enzyme.Angew Chem Int Ed Engl. 2021 May 10;60(20):11423-11429. doi: 10.1002/anie.202100969. Epub 2021 Apr 8. Angew Chem Int Ed Engl. 2021. PMID: 33661567 Free PMC article.
-
Biosynthesis of Fungal Natural Products Involving Two Separate Pathway Crosstalk.J Fungi (Basel). 2022 Mar 21;8(3):320. doi: 10.3390/jof8030320. J Fungi (Basel). 2022. PMID: 35330322 Free PMC article. Review.
-
Carbon-nitrogen bond formation to construct novel polyketide-indole hybrids from the indole-3-carbinol exposed culture of Daldinia eschscholzii.Synth Syst Biotechnol. 2022 Mar 19;7(2):750-755. doi: 10.1016/j.synbio.2022.02.004. eCollection 2022 Jun. Synth Syst Biotechnol. 2022. PMID: 35387230 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

