Triggering the expression of a silent gene cluster from genetically intractable bacteria results in scleric acid discovery
- PMID: 30746093
- PMCID: PMC6335953
- DOI: 10.1039/c8sc03814g
Triggering the expression of a silent gene cluster from genetically intractable bacteria results in scleric acid discovery
Abstract
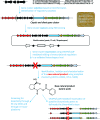
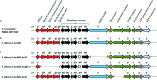
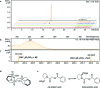
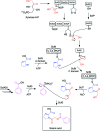
In this study, we report the rapid characterisation of a novel microbial natural product resulting from the rational derepression of a silent gene cluster. A conserved set of five regulatory genes was used as a query to search genomic databases and identify atypical biosynthetic gene clusters (BGCs). A 20-kb BGC from the genetically intractable Streptomyces sclerotialus bacterial strain was captured using yeast-based homologous recombination and introduced into validated heterologous hosts. CRISPR/Cas9-mediated genome editing was then employed to rationally inactivate the key transcriptional repressor and trigger production of an unprecedented class of hybrid natural products exemplified by (2-(benzoyloxy)acetyl)-l-proline, named scleric acid. Subsequent rounds of CRISPR/Cas9-mediated gene deletions afforded a selection of biosynthetic gene mutant strains which led to a plausible biosynthetic pathway for scleric acid assembly. Synthetic standards of scleric acid and a key biosynthetic intermediate were also prepared to confirm the chemical structures we proposed. The assembly of scleric acid involves two unique condensation reactions catalysed by a single NRPS module and an ATP-grasp enzyme that link a proline and a benzoyl residue to each end of a rare hydroxyethyl-ACP intermediate, respectively. Scleric acid was shown to exhibit moderate inhibition activity against Mycobacterium tuberculosis, as well as inhibition of the cancer-associated metabolic enzyme nicotinamide N-methyltransferase (NNMT).
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

