Evaluation of segmentation algorithms for optical coherence tomography images of ovarian tissue
- PMID: 30746391
- PMCID: PMC6350616
- DOI: 10.1117/1.JMI.6.1.014002
Evaluation of segmentation algorithms for optical coherence tomography images of ovarian tissue
Abstract
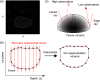
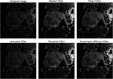
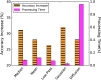
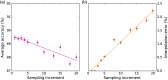
Ovarian cancer has the lowest survival rate among all gynecologic cancers predominantly due to late diagnosis. Early detection of ovarian cancer can increase 5-year survival rates from 40% up to 92%, yet no reliable early detection techniques exist. Optical coherence tomography (OCT) is an emerging technique that provides depth-resolved, high-resolution images of biological tissue in real-time and demonstrates great potential for imaging of ovarian tissue. Mouse models are crucial to quantitatively assess the diagnostic potential of OCT for ovarian cancer imaging; however, due to small organ size, the ovaries must first be separated from the image background using the process of segmentation. Manual segmentation is time-intensive, as OCT yields three-dimensional data. Furthermore, speckle noise complicates OCT images, frustrating many processing techniques. While much work has investigated noise-reduction and automated segmentation for retinal OCT imaging, little has considered the application to the ovaries, which exhibit higher variance and inhomogeneity than the retina. To address these challenges, we evaluate a set of algorithms to segment OCT images of mouse ovaries. We examine five preprocessing techniques and seven segmentation algorithms. While all preprocessing methods improve segmentation, Gaussian filtering is most effective, showing an improvement of . Of the segmentation algorithms, active contours performs best, segmenting with an accuracy of compared with manual segmentation. Even so, further optimization could lead to maximizing the performance for segmenting OCT images of the ovaries.
Keywords: image processing; image segmentation; optical coherence tomography; ovarian cancer.
Figures