Overriding FUS autoregulation in mice triggers gain-of-toxic dysfunctions in RNA metabolism and autophagy-lysosome axis
- PMID: 30747709
- PMCID: PMC6389288
- DOI: 10.7554/eLife.40811
Overriding FUS autoregulation in mice triggers gain-of-toxic dysfunctions in RNA metabolism and autophagy-lysosome axis
Abstract
Mutations in coding and non-coding regions of FUS cause amyotrophic lateral sclerosis (ALS). The latter mutations may exert toxicity by increasing FUS accumulation. We show here that broad expression within the nervous system of wild-type or either of two ALS-linked mutants of human FUS in mice produces progressive motor phenotypes accompanied by characteristic ALS-like pathology. FUS levels are autoregulated by a mechanism in which human FUS downregulates endogenous FUS at mRNA and protein levels. Increasing wild-type human FUS expression achieved by saturating this autoregulatory mechanism produces a rapidly progressive phenotype and dose-dependent lethality. Transcriptome analysis reveals mis-regulation of genes that are largely not observed upon FUS reduction. Likely mechanisms for FUS neurotoxicity include autophagy inhibition and defective RNA metabolism. Thus, our results reveal that overriding FUS autoregulation will trigger gain-of-function toxicity via altered autophagy-lysosome pathway and RNA metabolism function, highlighting a role for protein and RNA dyshomeostasis in FUS-mediated toxicity.
Keywords: FUS; RNA metabolism; amyotrophic lateral sclerosis (ALS); autophagy-lysosome; frontotemporal degeneration (FTD); homeostasis; mouse; neuroscience.
Conflict of interest statement
SL, SD, ST, WH, KL, HI, PP, ST, TT, JC, OP, NB, AB, AV, SS, MM, JH, DS, KK, GY, LT, MM, CS, GT, AL, CL, SD No competing interests declared, DC Reviewing editor, eLife
Figures


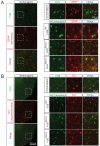



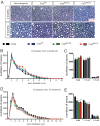
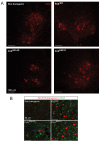

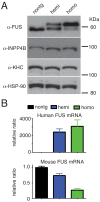

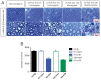
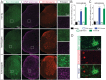



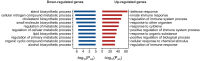

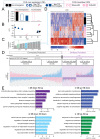

Comment in
-
Elevated FUS levels by overriding its autoregulation produce gain-of-toxicity properties that disrupt protein and RNA homeostasis.Autophagy. 2019 Sep;15(9):1665-1667. doi: 10.1080/15548627.2019.1633162. Epub 2019 Jun 23. Autophagy. 2019. PMID: 31230528 Free PMC article.
References
-
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochemical and Biophysical Research Communications. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. - DOI - PubMed
-
- Arnold ES, Ling SC, Huelga SC, Lagier-Tourenne C, Polymenidou M, Ditsworth D, Kordasiewicz HB, McAlonis-Downes M, Platoshyn O, Parone PA, Da Cruz S, Clutario KM, Swing D, Tessarollo L, Marsala M, Shaw CE, Yeo GW, Cleveland DW. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. PNAS. 2013;110:E736–E745. doi: 10.1073/pnas.1222809110. - DOI - PMC - PubMed
-
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

