Altered human oligodendrocyte heterogeneity in multiple sclerosis
- PMID: 30747918
- PMCID: PMC6544546
- DOI: 10.1038/s41586-019-0903-2
Altered human oligodendrocyte heterogeneity in multiple sclerosis
Abstract
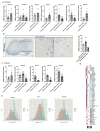
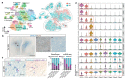
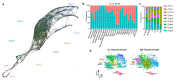
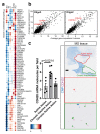
Oligodendrocyte pathology is increasingly implicated in neurodegenerative diseases as oligodendrocytes both myelinate and provide metabolic support to axons. In multiple sclerosis (MS), demyelination in the central nervous system thus leads to neurodegeneration, but the severity of MS between patients is very variable. Disability does not correlate well with the extent of demyelination1, which suggests that other factors contribute to this variability. One such factor may be oligodendrocyte heterogeneity. Not all oligodendrocytes are the same-those from the mouse spinal cord inherently produce longer myelin sheaths than those from the cortex2, and single-cell analysis of the mouse central nervous system identified further differences3,4. However, the extent of human oligodendrocyte heterogeneity and its possible contribution to MS pathology remain unknown. Here we performed single-nucleus RNA sequencing from white matter areas of post-mortem human brain from patients with MS and from unaffected controls. We identified subclusters of oligodendroglia in control human white matter, some with similarities to mouse, and defined new markers for these cell states. Notably, some subclusters were underrepresented in MS tissue, whereas others were more prevalent. These differences in mature oligodendrocyte subclusters may indicate different functional states of oligodendrocytes in MS lesions. We found similar changes in normal-appearing white matter, showing that MS is a more diffuse disease than its focal demyelination suggests. Our findings of an altered oligodendroglial heterogeneity in MS may be important for understanding disease progression and developing therapeutic approaches.
Conflict of interest statement
The authors declare competing financial interests. DM and IK are employees at F. Hoffmann – La Roche, Ltd.
Figures









Comment in
-
Oligodendrocyte studies reveal limits on remyelination.Nat Rev Neurol. 2019 Mar;15(3):125. doi: 10.1038/s41582-019-0149-5. Nat Rev Neurol. 2019. PMID: 30755723 No abstract available.
-
Time to revisit oligodendrocytes in multiple sclerosis.Nat Med. 2019 Mar;25(3):364-366. doi: 10.1038/s41591-019-0388-4. Nat Med. 2019. PMID: 30804516 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

