Using deep learning to identify translational research in genomic medicine beyond bench to bedside
- PMID: 30753477
- PMCID: PMC6367517
- DOI: 10.1093/database/baz010
Using deep learning to identify translational research in genomic medicine beyond bench to bedside
Abstract
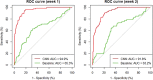
Tracking scientific research publications on the evaluation, utility and implementation of genomic applications is critical for the translation of basic research to impact clinical and population health. In this work, we utilize state-of-the-art machine learning approaches to identify translational research in genomics beyond bench to bedside from the biomedical literature. We apply the convolutional neural networks (CNNs) and support vector machines (SVMs) to the bench/bedside article classification on the weekly manual annotation data of the Public Health Genomics Knowledge Base database. Both classifiers employ salient features to determine the probability of curation-eligible publications, which can effectively reduce the workload of manual triage and curation process. We applied the CNNs and SVMs to an independent test set (n = 400), and the models achieved the F-measure of 0.80 and 0.74, respectively. We further tested the CNNs, which perform better results, on the routine annotation pipeline for 2 weeks and significantly reduced the effort and retrieved more appropriate research articles. Our approaches provide direct insight into the automated curation of genomic translational research beyond bench to bedside. The machine learning classifiers are found to be helpful for annotators to enhance the efficiency of manual curation.
Figures
Similar articles
-
Scaling up data curation using deep learning: An application to literature triage in genomic variation resources.PLoS Comput Biol. 2018 Aug 13;14(8):e1006390. doi: 10.1371/journal.pcbi.1006390. eCollection 2018 Aug. PLoS Comput Biol. 2018. PMID: 30102703 Free PMC article.
-
Comparing deep belief networks with support vector machines for classifying gene expression data from complex disorders.FEBS Open Bio. 2019 Jul;9(7):1232-1248. doi: 10.1002/2211-5463.12652. Epub 2019 Jun 7. FEBS Open Bio. 2019. PMID: 31074948 Free PMC article.
-
Horizon scanning for translational genomic research beyond bench to bedside.Genet Med. 2014 Jul;16(7):535-8. doi: 10.1038/gim.2013.184. Epub 2014 Jan 9. Genet Med. 2014. PMID: 24406461 Free PMC article.
-
Deep learning in medical image analysis: A third eye for doctors.J Stomatol Oral Maxillofac Surg. 2019 Sep;120(4):279-288. doi: 10.1016/j.jormas.2019.06.002. Epub 2019 Jun 26. J Stomatol Oral Maxillofac Surg. 2019. PMID: 31254638
-
Deep Learning-Based Algorithms in Screening of Diabetic Retinopathy: A Systematic Review of Diagnostic Performance.Ophthalmol Retina. 2019 Apr;3(4):294-304. doi: 10.1016/j.oret.2018.10.014. Epub 2018 Nov 3. Ophthalmol Retina. 2019. PMID: 31014679
Cited by
-
Methods for identifying biomedical translation: a systematic review.Am J Transl Res. 2022 Apr 15;14(4):2697-2708. eCollection 2022. Am J Transl Res. 2022. PMID: 35559386 Free PMC article. Review.
-
Artificial intelligence in clinical and translational science: Successes, challenges and opportunities.Clin Transl Sci. 2022 Feb;15(2):309-321. doi: 10.1111/cts.13175. Epub 2021 Oct 30. Clin Transl Sci. 2022. PMID: 34706145 Free PMC article. Review.
-
LitSuggest: a web-based system for literature recommendation and curation using machine learning.Nucleic Acids Res. 2021 Jul 2;49(W1):W352-W358. doi: 10.1093/nar/gkab326. Nucleic Acids Res. 2021. PMID: 33950204 Free PMC article.
References
-
- Khoury M.J., Gwinn M., Yoon P.W. et al. (2007) The continuum of translation research in genomic medicine: how can we accelerate the appropriate integration of human genome discoveries into health care and disease prevention. Genet. Med., 9, 665–674. - PubMed
-
- CDC Office of Public Health Genomics Genomic Tests and Family History by Levels of Evidence. https://phgkb.cdc.gov/PHGKB/topicStartPage.action Access date: July, 2018.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources