Criteria2Query: a natural language interface to clinical databases for cohort definition
- PMID: 30753493
- PMCID: PMC6402359
- DOI: 10.1093/jamia/ocy178
Criteria2Query: a natural language interface to clinical databases for cohort definition
Abstract
Objective: Cohort definition is a bottleneck for conducting clinical research and depends on subjective decisions by domain experts. Data-driven cohort definition is appealing but requires substantial knowledge of terminologies and clinical data models. Criteria2Query is a natural language interface that facilitates human-computer collaboration for cohort definition and execution using clinical databases.
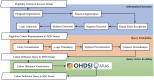
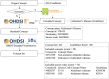
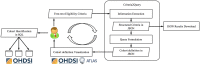
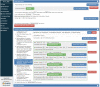
Materials and methods: Criteria2Query uses a hybrid information extraction pipeline combining machine learning and rule-based methods to systematically parse eligibility criteria text, transforms it first into a structured criteria representation and next into sharable and executable clinical data queries represented as SQL queries conforming to the OMOP Common Data Model. Users can interactively review, refine, and execute queries in the ATLAS web application. To test effectiveness, we evaluated 125 criteria across different disease domains from ClinicalTrials.gov and 52 user-entered criteria. We evaluated F1 score and accuracy against 2 domain experts and calculated the average computation time for fully automated query formulation. We conducted an anonymous survey evaluating usability.
Results: Criteria2Query achieved 0.795 and 0.805 F1 score for entity recognition and relation extraction, respectively. Accuracies for negation detection, logic detection, entity normalization, and attribute normalization were 0.984, 0.864, 0.514 and 0.793, respectively. Fully automatic query formulation took 1.22 seconds/criterion. More than 80% (11+ of 13) of users would use Criteria2Query in their future cohort definition tasks.
Conclusions: We contribute a novel natural language interface to clinical databases. It is open source and supports fully automated and interactive modes for autonomous data-driven cohort definition by researchers with minimal human effort. We demonstrate its promising user friendliness and usability.
Keywords: cohort definition; common data model; natural language interfaces to database; natural language processing.
© The Author(s) 2019. Published by Oxford University Press on behalf of the American Medical Informatics Association.
Figures


Similar articles
-
EliIE: An open-source information extraction system for clinical trial eligibility criteria.J Am Med Inform Assoc. 2017 Nov 1;24(6):1062-1071. doi: 10.1093/jamia/ocx019. J Am Med Inform Assoc. 2017. PMID: 28379377 Free PMC article.
-
Combining human and machine intelligence for clinical trial eligibility querying.J Am Med Inform Assoc. 2022 Jun 14;29(7):1161-1171. doi: 10.1093/jamia/ocac051. J Am Med Inform Assoc. 2022. PMID: 35426943 Free PMC article.
-
Evaluation of Criteria2Query: Towards Augmented Intelligence for Cohort Identification.Stud Health Technol Inform. 2022 Jun 6;290:297-300. doi: 10.3233/SHTI220082. Stud Health Technol Inform. 2022. PMID: 35673021
-
Formal representation of eligibility criteria: a literature review.J Biomed Inform. 2010 Jun;43(3):451-67. doi: 10.1016/j.jbi.2009.12.004. Epub 2009 Dec 23. J Biomed Inform. 2010. PMID: 20034594 Free PMC article. Review.
-
LinkHub: a Semantic Web system that facilitates cross-database queries and information retrieval in proteomics.BMC Bioinformatics. 2007 May 9;8 Suppl 3(Suppl 3):S5. doi: 10.1186/1471-2105-8-S3-S5. BMC Bioinformatics. 2007. PMID: 17493288 Free PMC article. Review.
Cited by
-
Named Entity Recognition and Normalization for Alzheimer's Disease Eligibility Criteria.Proc (IEEE Int Conf Healthc Inform). 2023 Jun;2023:558-564. doi: 10.1109/ichi57859.2023.00100. Epub 2023 Dec 11. Proc (IEEE Int Conf Healthc Inform). 2023. PMID: 38283164 Free PMC article.
-
How can natural language processing help model informed drug development?: a review.JAMIA Open. 2022 Jun 11;5(2):ooac043. doi: 10.1093/jamiaopen/ooac043. eCollection 2022 Jul. JAMIA Open. 2022. PMID: 35702625 Free PMC article. Review.
-
The IMPACT framework and implementation for accessible in silico clinical phenotyping in the digital era.NPJ Digit Med. 2023 Jul 21;6(1):132. doi: 10.1038/s41746-023-00878-9. NPJ Digit Med. 2023. PMID: 37479735 Free PMC article. Review.
-
A guide to artificial intelligence for cancer researchers.Nat Rev Cancer. 2024 Jun;24(6):427-441. doi: 10.1038/s41568-024-00694-7. Epub 2024 May 16. Nat Rev Cancer. 2024. PMID: 38755439 Review.
-
Prototypical Clinical Trial Registry Based on Fast Healthcare Interoperability Resources (FHIR): Design and Implementation Study.JMIR Med Inform. 2021 Jan 12;9(1):e20470. doi: 10.2196/20470. JMIR Med Inform. 2021. PMID: 33433393 Free PMC article.
References
-
- Häyrinen K, Saranto K, Nykänen P.. Definition, structure, content, use and impacts of electronic health records: a review of the research literature. Int J Med Inf 2008; 77: 291–304. - PubMed
-
- Musen MA, Rohn JA, Fagan LM, et al. Knowledge engineering for a clinical trial advice system: uncovering errors in protocol specification. Bull Cancer 1987; 74: 291–6. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

