The E3-ligases SCFPpa and APC/CCdh1 co-operate to regulate CENP-ACID expression across the cell cycle
- PMID: 30753559
- PMCID: PMC6468245
- DOI: 10.1093/nar/gkz060
The E3-ligases SCFPpa and APC/CCdh1 co-operate to regulate CENP-ACID expression across the cell cycle
Abstract
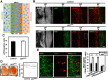
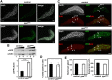
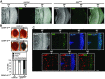
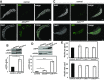
Centromere identity is determined by the specific deposition of CENP-A, a histone H3 variant localizing exclusively at centromeres. Increased CENP-A expression, which is a frequent event in cancer, causes mislocalization, ectopic kinetochore assembly and genomic instability. Proteolysis regulates CENP-A expression and prevents its misincorporation across chromatin. How proteolysis restricts CENP-A localization to centromeres is not well understood. Here we report that, in Drosophila, CENP-ACID expression levels are regulated throughout the cell cycle by the combined action of SCFPpa and APC/CCdh1. We show that SCFPpa regulates CENP-ACID expression in G1 and, importantly, in S-phase preventing its promiscuous incorporation across chromatin during replication. In G1, CENP-ACID expression is also regulated by APC/CCdh1. We also show that Cal1, the specific chaperone that deposits CENP-ACID at centromeres, protects CENP-ACID from SCFPpa-mediated degradation but not from APC/CCdh1-mediated degradation. These results suggest that, whereas SCFPpa targets the fraction of CENP-ACID that is not in complex with Cal1, APC/CCdh1 mediates also degradation of the Cal1-CENP-ACID complex and, thus, likely contributes to the regulation of centromeric CENP-ACID deposition.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Phosphorylation of Drosophila CENP-A on serine 20 regulates protein turn-over and centromere-specific loading.Nucleic Acids Res. 2019 Nov 18;47(20):10754-10770. doi: 10.1093/nar/gkz809. Nucleic Acids Res. 2019. PMID: 31535131 Free PMC article.
-
Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres.Nucleic Acids Res. 2006;34(21):6247-55. doi: 10.1093/nar/gkl902. Epub 2006 Nov 7. Nucleic Acids Res. 2006. PMID: 17090596 Free PMC article.
-
Establishment of Centromeric Chromatin by the CENP-A Assembly Factor CAL1 Requires FACT-Mediated Transcription.Dev Cell. 2015 Jul 6;34(1):73-84. doi: 10.1016/j.devcel.2015.05.012. Dev Cell. 2015. PMID: 26151904 Free PMC article.
-
Orchestrating the Specific Assembly of Centromeric Nucleosomes.Prog Mol Subcell Biol. 2017;56:165-192. doi: 10.1007/978-3-319-58592-5_7. Prog Mol Subcell Biol. 2017. PMID: 28840237 Free PMC article. Review.
-
Centromeric and ectopic assembly of CENP-A chromatin in health and cancer: old marks and new tracks.Nucleic Acids Res. 2019 Feb 20;47(3):1051-1069. doi: 10.1093/nar/gky1298. Nucleic Acids Res. 2019. PMID: 30590707 Free PMC article. Review.
Cited by
-
APC/C ubiquitin ligase: Functions and mechanisms in tumorigenesis.Semin Cancer Biol. 2020 Dec;67(Pt 2):80-91. doi: 10.1016/j.semcancer.2020.03.001. Epub 2020 Mar 9. Semin Cancer Biol. 2020. PMID: 32165320 Free PMC article. Review.
-
Recent insights into mechanisms preventing ectopic centromere formation.Open Biol. 2021 Sep;11(9):210189. doi: 10.1098/rsob.210189. Epub 2021 Sep 8. Open Biol. 2021. PMID: 34493071 Free PMC article. Review.
-
Minimal Cylinder Analysis Reveals the Mechanical Properties of Oncogenic Nucleosomes.Biophys J. 2020 May 5;118(9):2309-2318. doi: 10.1016/j.bpj.2020.01.042. Epub 2020 Feb 12. Biophys J. 2020. PMID: 32097625 Free PMC article.
-
Reduce, Retain, Recycle: Mechanisms for Promoting Histone Protein Degradation versus Stability and Retention.Mol Cell Biol. 2021 May 21;41(6):e0000721. doi: 10.1128/MCB.00007-21. Epub 2021 May 21. Mol Cell Biol. 2021. PMID: 33753462 Free PMC article. Review.
-
Localization of Drosophila CENP-A to non-centromeric sites depends on the NuRD complex.Nucleic Acids Res. 2019 Dec 16;47(22):11589-11608. doi: 10.1093/nar/gkz962. Nucleic Acids Res. 2019. PMID: 31713634 Free PMC article.
References
-
- Malik H.S., Henikoff S.. Major evolutionay transitions in centromere complexity. Cell. 2009; 138:1067–1082. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

