Signals of Variation in Human Mutation Rate at Multiple Levels of Sequence Context
- PMID: 30753705
- PMCID: PMC6501879
- DOI: 10.1093/molbev/msz023
Signals of Variation in Human Mutation Rate at Multiple Levels of Sequence Context
Abstract
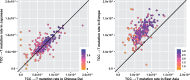
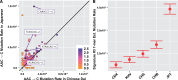
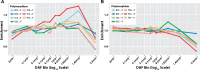
Our understanding of the human mutation rate helps us build evolutionary models and interpret patterns of genetic variation observed in human populations. Recent work indicates that the frequencies of specific polymorphism types have been elevated in Europe, and that many more, subtler signatures of global polymorphism variation may yet remain unidentified. Here, we present an analysis of the 1000 Genomes Project supported by analysis in the Simons Genome Diversity Panel, suggesting additional putative signatures of mutation rate variation across populations and the extent to which they are shaped by local sequence context. First, we compiled a list of the most significantly variable polymorphism types in a cross-continental statistical test. Clustering polymorphisms together, we observe three sets that showed distinct shared patterns of relative enrichment among ancestral populations, and we characterize each one of these putative "signatures" of polymorphism variation. For three of these signatures, we found that a single flanking base pair of sequence context was sufficient to determine the majority of enrichment or depletion of a polymorphism type. However, local genetic context up to 2-3 bp away contributes additional variability and may help to interpret a previously noted enrichment of certain polymorphism types in some East Asian groups. Moreover, considering broader local genetic context highlights patterns of polymorphism variation, which were not captured by previous approaches. Building our understanding of mutation rate in this way can help us to construct more accurate evolutionary models and better understand the mechanisms that underlie genetic change.
Keywords: mutation rate; sequence context models; statistical genetics.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

