Transcriptome Analysis Reveals New Insights into the Bacterial Wilt Resistance Mechanism Mediated by Silicon in Tomato
- PMID: 30754671
- PMCID: PMC6387441
- DOI: 10.3390/ijms20030761
Transcriptome Analysis Reveals New Insights into the Bacterial Wilt Resistance Mechanism Mediated by Silicon in Tomato
Abstract
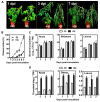
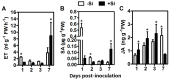
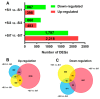
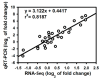
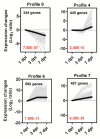
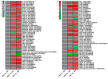
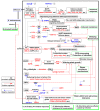
Bacterial wilt is a devastating disease of tomato caused by soilborne pathogenic bacterium Ralstonia solanacearum. Previous studies found that silicon (Si) can increase tomato resistance against R. solanacearum, but the exact molecular mechanism remains unclear. RNA sequencing (RNA-Seq) technology was used to investigate the dynamic changes of root transcriptome profiles between Si-treated (+Si) and untreated (-Si) tomato plants at 1, 3, and 7 days post-inoculation with R. solanacearum. The contents of salicylic acid (SA), ethylene (ET), and jasmonic acid (JA) and the activity of defense-related enzymes in roots of tomato in different treatments were also determined. The burst of ET production in roots was delayed, and SA and JA contents were altered in Si treatment. The transcriptional response to R. solanacearum infection of the +Si plants was quicker than that of the untreated plants. The expression levels of differentially-expressed genes involved in pathogen-associated molecular pattern-triggered immunity (PTI), oxidation resistance, and water-deficit stress tolerance were upregulated in the Si-treated plants. Multiple hormone-related genes were differentially expressed in the Si-treated plants. Si-mediated resistance involves mechanisms other than SA- and JA/ET-mediated stress responses. We propose that Si-mediated tomato resistance to R. solanacearum is associated with activated PTI-related responses and enhanced disease resistance and tolerance via several signaling pathways. Such pathways are mediated by multiple hormones (e.g., SA, JA, ET, and auxin), leading to diminished adverse effects (e.g., senescence, water-deficit, salinity and oxidative stress) normally caused by R. solanacearum infection. This finding will provide an important basis to further characterize the role of Si in enhancing plant resistance against biotic stress.
Keywords: Ralstonia solanacearum; induced resistance; silicon; tomato; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Dannon E.A., Wydra K. Interaction between silicon amendment, bacterial wilt development and phenotype of Ralstonia solanacearum in tomato genotypes. Physiol. Mol. Plant Pathol. 2004;64:233–243. doi: 10.1016/j.pmpp.2004.09.006. - DOI
-
- Chen Y.T., Liu M., Wang L., Lin W.P., Fan X.Y., Cai K.Z. Proteomic characterization of silicon-mediated resistance against Ralstonia solanacearum in tomato. Plant Soil. 2015;387:425–440. doi: 10.1007/s11104-014-2293-4. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

