Transcriptome Analysis of Diurnal Gene Expression in Chinese Cabbage
- PMID: 30754711
- PMCID: PMC6409912
- DOI: 10.3390/genes10020130
Transcriptome Analysis of Diurnal Gene Expression in Chinese Cabbage
Abstract
Plants have developed timing mechanisms that enable them to maintain synchrony with daily environmental events. These timing mechanisms, i.e., circadian clocks, include transcriptional/translational feedback loops that drive 24 h transcriptional rhythms, which underlie oscillations in protein abundance, thus mediating circadian rhythms of behavior, physiology, and metabolism. Circadian clock genes have been investigated in the diploid model plant Arabidopsis thaliana. Crop plants with polyploid genomes-such as Brassica species-have multiple copies of some clock-related genes. Over the last decade, numerous studies have been aimed at identifying and understanding the function of paralogous genes with conserved sequences, or those that diverged during evolution. Brassicarapa's triplicate genomes retain sequence-level collinearity with Arabidopsis. In this study, we used RNA sequencing (RNAseq) to profile the diurnal transcriptome of Brassicarapa seedlings. We identified candidate paralogs of circadian clock-related genes and assessed their expression levels. These genes and their related traits that modulate the diurnal rhythm of gene expression contribute to the adaptation of crop cultivars. Our findings will contribute to the mechanistic study of circadian clock regulation inherent in polyploidy genome crops, which differ from those of model plants, and thus will be useful for future breeding studies using clock genes.
Keywords: Arabidopsis; Brassica rapa; circadian-related gene; polyploid genome; transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

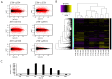
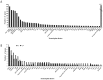

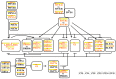
Similar articles
-
Expansion of the circadian transcriptome in Brassica rapa and genome-wide diversification of paralog expression patterns.Elife. 2020 Sep 30;9:e58993. doi: 10.7554/eLife.58993. Elife. 2020. PMID: 32996462 Free PMC article.
-
Comparative mapping, genomic structure, and expression analysis of eight pseudo-response regulator genes in Brassica rapa.Mol Genet Genomics. 2012 May;287(5):373-88. doi: 10.1007/s00438-012-0682-z. Epub 2012 Apr 1. Mol Genet Genomics. 2012. PMID: 22466714
-
Interspecific analysis of diurnal gene regulation in panicoid grasses identifies known and novel regulatory motifs.BMC Genomics. 2020 Jun 25;21(1):428. doi: 10.1186/s12864-020-06824-3. BMC Genomics. 2020. PMID: 32586356 Free PMC article.
-
Physiological significance of the plant circadian clock in natural field conditions.Plant Cell Environ. 2012 Oct;35(10):1729-41. doi: 10.1111/j.1365-3040.2012.02555.x. Epub 2012 Jul 18. Plant Cell Environ. 2012. PMID: 22681566 Review.
-
Circadian clock signaling in Arabidopsis thaliana: from gene expression to physiology and development.Int J Dev Biol. 2005;49(5-6):491-500. doi: 10.1387/ijdb.041968pm. Int J Dev Biol. 2005. PMID: 16096959 Review.
Cited by
-
The BrGI Circadian Clock Gene Is Involved in the Regulation of Glucosinolates in Chinese Cabbage.Genes (Basel). 2021 Oct 22;12(11):1664. doi: 10.3390/genes12111664. Genes (Basel). 2021. PMID: 34828270 Free PMC article.
-
Circadian regulation of the transcriptome in a complex polyploid crop.PLoS Biol. 2022 Oct 13;20(10):e3001802. doi: 10.1371/journal.pbio.3001802. eCollection 2022 Oct. PLoS Biol. 2022. PMID: 36227835 Free PMC article.
-
Alternative polyadenylated mRNAs behave as asynchronous rhythmic transcription in Arabidopsis.RNA Biol. 2021 Dec;18(12):2594-2604. doi: 10.1080/15476286.2021.1933732. Epub 2021 Jun 7. RNA Biol. 2021. PMID: 34036876 Free PMC article.
-
Identification and Characterization of PSEUDO-RESPONSE REGULATOR (PRR) 1a and 1b Genes by CRISPR/Cas9-Targeted Mutagenesis in Chinese Cabbage (Brassica rapa L.).Int J Mol Sci. 2022 Jun 23;23(13):6963. doi: 10.3390/ijms23136963. Int J Mol Sci. 2022. PMID: 35806003 Free PMC article.
-
QTL Mapping and Diurnal Transcriptome Analysis Identify Candidate Genes Regulating Brassica napus Flowering Time.Int J Mol Sci. 2021 Jul 15;22(14):7559. doi: 10.3390/ijms22147559. Int J Mol Sci. 2021. PMID: 34299178 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

