Acidification of Tumor at Stromal Boundaries Drives Transcriptome Alterations Associated with Aggressive Phenotypes
- PMID: 30755444
- PMCID: PMC6467770
- DOI: 10.1158/0008-5472.CAN-18-1604
Acidification of Tumor at Stromal Boundaries Drives Transcriptome Alterations Associated with Aggressive Phenotypes
Abstract
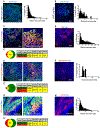
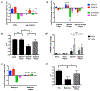
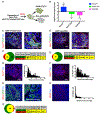
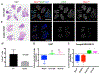
Acidosis is a fundamental feature of the tumor microenvironment, which directly regulates tumor cell invasion by affecting immune cell function, clonal cell evolution, and drug resistance. Despite the important association of tumor microenvironment acidosis with tumor cell invasion, relatively little is known regarding which areas within a tumor are acidic and how acidosis influences gene expression to promote invasion. Here, we injected a labeled pH-responsive peptide to mark acidic regions within tumors. Surprisingly, acidic regions were not restricted to hypoxic areas and overlapped with highly proliferative, invasive regions at the tumor-stroma interface, which were marked by increased expression of matrix metalloproteinases and degradation of the basement membrane. RNA-seq analysis of cells exposed to low pH conditions revealed a general rewiring of the transcriptome that involved RNA splicing and enriched for targets of RNA binding proteins with specificity for AU-rich motifs. Alternative splicing of Mena and CD44, which play important isoform-specific roles in metastasis and drug resistance, respectively, was sensitive to histone acetylation status. Strikingly, this program of alternative splicing was reversed in vitro and in vivo through neutralization experiments that mitigated acidic conditions. These findings highlight a previously underappreciated role for localized acidification of tumor microenvironment in the expression of an alternative splicing-dependent tumor invasion program. SIGNIFICANCE: This study expands our understanding of acidosis within the tumor microenvironment and indicates that acidosis induces potentially therapeutically actionable changes to alternative splicing.
©2019 American Association for Cancer Research.
Conflict of interest statement
Conflict of interest:
The authors declare no potential conflicts of interest.
Figures



References
-
- Andreucci E, Peppicelli S, Carta F, Brisotto G, Biscontin E, Ruzzolini J, et al. Carbonic anhydrase IX inhibition affects viability of cancer cells adapted to extracellular acidosis. J Mol Med. Journal of Molecular Medicine; 2017;95:1341–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

