Hierarchical Transcription Factor and Chromatin Binding Network for Wood Formation in Black Cottonwood (Populus trichocarpa)
- PMID: 30755461
- PMCID: PMC6482634
- DOI: 10.1105/tpc.18.00620
Hierarchical Transcription Factor and Chromatin Binding Network for Wood Formation in Black Cottonwood (Populus trichocarpa)
Abstract
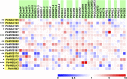
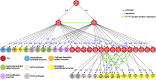
Wood remains the world's most abundant and renewable resource for timber and pulp and is an alternative to fossil fuels. Understanding the molecular regulation of wood formation can advance the engineering of wood for more efficient material and energy productions. We integrated a black cottonwood (Populus trichocarpa) wood-forming cell system with quantitative transcriptomics and chromatin binding assays to construct a transcriptional regulatory network (TRN) directed by a key transcription factor (TF), PtrSND1-B1 (secondary wall-associated NAC-domain protein). The network consists of four layers of TF-target gene interactions with quantitative regulatory effects, describing the specificity of how the regulation is transduced through these interactions to activate cell wall genes (effector genes) for wood formation. PtrSND1-B1 directs 57 TF-DNA interactions through 17 TFs transregulating 27 effector genes. Of the 57 interactions, 55 are novel. We tested 42 of these 57 interactions in 30 genotypes of transgenic P. trichocarpa and verified that ∼90% of the tested interactions function in vivo. The TRN reveals common transregulatory targets for distinct TFs, leading to the discovery of nine TF protein complexes (dimers and trimers) implicated in regulating the biosynthesis of specific types of lignin. Our work suggests that wood formation may involve regulatory homeostasis determined by combinations of TF-DNA and TF-TF (protein-protein) regulations.
© 2019 American Society of Plant Biologists. All rights reserved.
Figures









References
-
- Albersheim P., Darvill A., Roberts K., Sederoff R., Staehelin A. (2011). Cell walls and plant anatomy. In Albersheim P, Darvill A, Roberts K, Sederoff R, Staehelin A, eds, Plant Cell Walls: From Chemistry to Biology, Garland Science, New York, pp 1–42.
-
- Berthet S., Demont-Caulet N., Pollet B., Bidzinski P., Cézard L., Le Bris P., Borrega N., Hervé J., Blondet E., Balzergue S., Lapierre C., Jouanin L. (2011). Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137. - PMC - PubMed
-
- Bhardwaj N., Kim P.M., Gerstein M.B. (2010). Rewiring of transcriptional regulatory networks: Hierarchy, rather than connectivity, better reflects the importance of regulators. Sci. Signal. 3: ra79. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

