A MYB Triad Controls Primary and Phenylpropanoid Metabolites for Pollen Coat Patterning
- PMID: 30755473
- PMCID: PMC6501115
- DOI: 10.1104/pp.19.00009
A MYB Triad Controls Primary and Phenylpropanoid Metabolites for Pollen Coat Patterning
Abstract
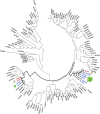
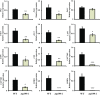
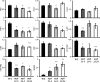
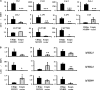
The pollen wall is a complex, durable structure essential for plant reproduction. A substantial portion of phenylpropanoids (e.g. flavonols) produced by pollen grain tapetal cells are deposited in the pollen wall. Transcriptional regulation of pollen wall formation has been studied extensively, and a specific regulatory mechanism for Arabidopsis (Arabidopsis thaliana) pollen flavonol biosynthesis has been postulated. Here, metabolome and transcriptome analyses of anthers from mutant and overexpression genotypes revealed that Arabidopsis MYB99, a putative ortholog of the petunia (Petunia hybrida) floral scent regulator ODORANT1 (ODO1), controls the exclusive production of tapetum diglycosylated flavonols and hydroxycinnamic acid amides. We discovered that MYB99 acts in a regulatory triad with MYB21 and MYB24, orthologs of emission of benzenoids I and II, which together with ODO1 coregulate petunia scent biosynthesis genes. Furthermore, promoter-activation assays showed that MYB99 directs precursor supply from the Calvin cycle and oxidative pentose-phosphate pathway in primary metabolism to phenylpropanoid biosynthesis by controlling TRANSKETOLASE2 expression. We provide a model depicting the relationship between the Arabidopsis MYB triad and structural genes from primary and phenylpropanoid metabolism and compare this mechanism with petunia scent control. The discovery of orthologous protein triads producing related secondary metabolites suggests that analogous regulatory modules exist in other plants and act to regulate various branches of the intricate phenylpropanoid pathway.
© 2019 American Society of Plant Biologists. All Rights Reserved.
Figures




References
-
- Aharoni A, Galili G (2011) Metabolic engineering of the plant primary-secondary metabolism interface. Curr Opin Biotechnol 22: 239–244 - PubMed
-
- Ahlers F, Lambert J, Wiermann R (1999) Structural elements of sporopollenin from the pollen of Torreya californica Torr. (Gymnospermae): Using the H-1-NMR technique. Z Naturforsch C 54: 492–495
-
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

