A Genome-Wide Analysis of Adhesion in Caulobacter crescentus Identifies New Regulatory and Biosynthetic Components for Holdfast Assembly
- PMID: 30755507
- PMCID: PMC6372794
- DOI: 10.1128/mBio.02273-18
A Genome-Wide Analysis of Adhesion in Caulobacter crescentus Identifies New Regulatory and Biosynthetic Components for Holdfast Assembly
Abstract
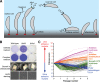
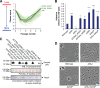
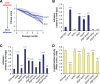
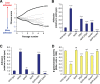
Due to their intimate physical interactions with the environment, surface polysaccharides are critical determinants of fitness for bacteria. Caulobacter crescentus produces a specialized structure at one of its cell poles called the holdfast that enables attachment to surfaces. Previous studies have shown that the holdfast is composed of carbohydrate-based material and identified a number of genes required for holdfast development. However, incomplete information about its chemical structure, biosynthetic genes, and regulatory principles has limited progress in understanding the mechanism of holdfast synthesis. We leveraged the adhesive properties of the holdfast to perform a saturating screen for genes affecting attachment to cheesecloth over a multiday time course. Using similarities in the temporal profiles of mutants in a transposon library, we defined discrete clusters of genes with related effects on cheesecloth colonization. Holdfast synthesis, flagellar motility, type IV pilus assembly, and smooth lipopolysaccharide (SLPS) production represented key classes of adhesion determinants. Examining these clusters in detail allowed us to predict and experimentally define the functions of multiple uncharacterized genes in both the holdfast and SLPS pathways. In addition, we showed that the pilus and the flagellum control holdfast synthesis separately by modulating the holdfast inhibitor hfiA. This report defines a set of genes contributing to adhesion that includes newly discovered genes required for holdfast biosynthesis and attachment. Our data provide evidence that the holdfast contains a complex polysaccharide with at least four monosaccharides in the repeating unit and underscore the central role of cell polarity in mediating attachment of C. crescentus to surfaces.IMPORTANCE Bacteria routinely encounter biotic and abiotic materials in their surrounding environments, and they often enlist specific behavioral programs to colonize these materials. Adhesion is an early step in colonizing a surface. Caulobacter crescentus produces a structure called the holdfast which allows this organism to attach to and colonize surfaces. To understand how the holdfast is produced, we performed a genome-wide search for genes that contribute to adhesion by selecting for mutants that could not attach to cheesecloth. We discovered complex interactions between genes that mediate surface contact and genes that contribute to holdfast development. Our genetic selection identified what likely represents a comprehensive set of genes required to generate a holdfast, laying the groundwork for a detailed characterization of the enzymes that build this specialized adhesin.
Keywords: BarSeq; Caulobacter; adhesion; holdfast; pilus; polysaccharide.
Copyright © 2019 Hershey et al.
Figures
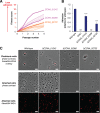
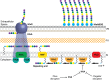
Similar articles
-
Flagellar Perturbations Activate Adhesion through Two Distinct Pathways in Caulobacter crescentus.mBio. 2021 Feb 9;12(1):e03266-20. doi: 10.1128/mBio.03266-20. mBio. 2021. PMID: 33563824 Free PMC article.
-
Role of Caulobacter Cell Surface Structures in Colonization of the Air-Liquid Interface.J Bacteriol. 2019 Aug 22;201(18):e00064-19. doi: 10.1128/JB.00064-19. Print 2019 Sep 15. J Bacteriol. 2019. PMID: 31010900 Free PMC article.
-
Composition of the Holdfast Polysaccharide from Caulobacter crescentus.J Bacteriol. 2019 Aug 8;201(17):e00276-19. doi: 10.1128/JB.00276-19. Print 2019 Sep 1. J Bacteriol. 2019. PMID: 31209074 Free PMC article.
-
Scaling down for a broader understanding of underwater adhesives - a case for the Caulobacter crescentus holdfast.Soft Matter. 2016 Nov 16;12(45):9132-9141. doi: 10.1039/c6sm02163h. Soft Matter. 2016. PMID: 27812588 Review.
-
Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus.Adv Microb Physiol. 2009;54:1-101. doi: 10.1016/S0065-2911(08)00001-5. Adv Microb Physiol. 2009. PMID: 18929067 Free PMC article. Review.
Cited by
-
An exopolysaccharide pathway from a freshwater Sphingomonas isolate.J Bacteriol. 2024 Aug 22;206(8):e0016924. doi: 10.1128/jb.00169-24. Epub 2024 Jul 15. J Bacteriol. 2024. PMID: 39007563 Free PMC article.
-
The HmrABCX pathway regulates the transition between motile and sessile lifestyles in Caulobacter crescentus by a HfiA-independent mechanism.bioRxiv [Preprint]. 2023 Dec 14:2023.12.13.571505. doi: 10.1101/2023.12.13.571505. bioRxiv. 2023. Update in: mBio. 2024 Oct 16;15(10):e0100224. doi: 10.1128/mbio.01002-24. PMID: 38168291 Free PMC article. Updated. Preprint.
-
Genome-wide identification of Pseudomonas syringae genes required for fitness during colonization of the leaf surface and apoplast.Proc Natl Acad Sci U S A. 2019 Sep 17;116(38):18900-18910. doi: 10.1073/pnas.1908858116. Epub 2019 Sep 4. Proc Natl Acad Sci U S A. 2019. PMID: 31484768 Free PMC article.
-
HfaE Is a Component of the Holdfast Anchor Complex That Tethers the Holdfast Adhesin to the Cell Envelope.J Bacteriol. 2022 Nov 15;204(11):e0027322. doi: 10.1128/jb.00273-22. Epub 2022 Sep 27. J Bacteriol. 2022. PMID: 36165621 Free PMC article.
-
Sticky decisions: The multilayered regulation of adhesin production by bacteria.PLoS Genet. 2023 Mar 2;19(3):e1010648. doi: 10.1371/journal.pgen.1010648. eCollection 2023 Mar. PLoS Genet. 2023. PMID: 36862629 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

