A Highly Polymorphic Receptor Governs Many Distinct Self-Recognition Types within the Myxococcales Order
- PMID: 30755513
- PMCID: PMC6372800
- DOI: 10.1128/mBio.02751-18
A Highly Polymorphic Receptor Governs Many Distinct Self-Recognition Types within the Myxococcales Order
Abstract
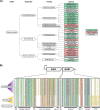
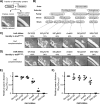
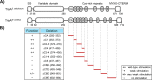
Self-recognition underlies sociality in many group-living organisms. In bacteria, cells use various strategies to recognize kin to form social groups and, in some cases, to transition into multicellular life. One strategy relies on a single genetic locus that encodes a variable phenotypic tag ("greenbeard") for recognizing other tag bearers. Previously, we discovered a polymorphic cell surface receptor called TraA that directs self-identification through homotypic interactions in the social bacterium Myxococcus xanthus Recognition by TraA leads to cellular resource sharing in a process called outer membrane exchange (OME). A second gene in the traA operon, traB, is also required for OME but is not involved in recognition. Our prior studies of TraA identified only six recognition groups among closely related M. xanthus isolates. Here we hypothesize that the number of traA polymorphisms and, consequently, the diversity of recognition in wild isolates are much greater. To test this hypothesis, we expand the scope of TraA characterization to the order Myxococcales From genomic sequences within the three suborders of Myxococcales, we identified 90 traA orthologs. Sequence analyses and functional characterization of traAB loci suggest that OME is well maintained among diverse myxobacterial taxonomic groups. Importantly, TraA orthologs are highly polymorphic within their variable domain, the region that confers selectivity in self-recognition. We experimentally defined 10 distinct recognition groups and, based on phylogenetic and experimental analyses, predicted >60 recognition groups among the 90 traA alleles. Taken together, our findings revealed a widespread greenbeard locus that mediates the diversity of self-recognition across the order MyxococcalesIMPORTANCE Many biological species distinguish self from nonself by using different mechanisms. Higher animals recognize close kin via complex processes that often involve the five senses, cognition, and learning, whereas some microbes achieve self-recognition simply through the activity of a single genetic locus. Here we describe a single locus, traA, in myxobacteria that governs cell-cell recognition within natural populations. We found that traA is widespread across the order Myxococcales TraA is highly polymorphic among diverse myxobacterial isolates, and such polymorphisms determine selectivity in self-recognition. Through bioinformatic and experimental analyses, we showed that traA governs many distinct recognition groups within Myxococcales This report provides an example in which a single locus influences social recognition across a wide phylogenetic range of natural populations.
Keywords: cell surface; cellular transfer; kin recognition; myxobacteria; polymorphism.
Copyright © 2019 Cao et al.
Figures



Similar articles
-
Kin recognition and outer membrane exchange (OME) in myxobacteria.Curr Opin Microbiol. 2020 Aug;56:81-88. doi: 10.1016/j.mib.2020.07.003. Epub 2020 Aug 20. Curr Opin Microbiol. 2020. PMID: 32828979 Free PMC article. Review.
-
Self-identity barcodes encoded by six expansive polymorphic toxin families discriminate kin in myxobacteria.Proc Natl Acad Sci U S A. 2019 Dec 3;116(49):24808-24818. doi: 10.1073/pnas.1912556116. Epub 2019 Nov 19. Proc Natl Acad Sci U S A. 2019. PMID: 31744876 Free PMC article.
-
Molecular recognition in myxobacterial outer membrane exchange: functional, social and evolutionary implications.Mol Microbiol. 2014 Jan;91(2):209-20. doi: 10.1111/mmi.12450. Epub 2013 Nov 21. Mol Microbiol. 2014. PMID: 24261719 Free PMC article. Review.
-
Sibling Rivalry in Myxococcus xanthus Is Mediated by Kin Recognition and a Polyploid Prophage.J Bacteriol. 2016 Jan 19;198(6):994-1004. doi: 10.1128/JB.00964-15. J Bacteriol. 2016. PMID: 26787762 Free PMC article.
-
Self-identity reprogrammed by a single residue switch in a cell surface receptor of a social bacterium.Proc Natl Acad Sci U S A. 2017 Apr 4;114(14):3732-3737. doi: 10.1073/pnas.1700315114. Epub 2017 Mar 20. Proc Natl Acad Sci U S A. 2017. PMID: 28320967 Free PMC article.
Cited by
-
A Polymorphic Gene within the Mycobacterium smegmatis esx1 Locus Determines Mycobacterial Self-Identity and Conjugal Compatibility.mBio. 2022 Apr 26;13(2):e0021322. doi: 10.1128/mbio.00213-22. Epub 2022 Mar 17. mBio. 2022. PMID: 35297678 Free PMC article.
-
New binding specificities evolve via point mutation in an invertebrate allorecognition gene.iScience. 2021 Jul 1;24(7):102811. doi: 10.1016/j.isci.2021.102811. eCollection 2021 Jul 23. iScience. 2021. PMID: 34296075 Free PMC article.
-
Two reasons to kill: predation and kin discrimination in myxobacteria.Microbiology (Reading). 2023 Jul;169(7):001372. doi: 10.1099/mic.0.001372. Microbiology (Reading). 2023. PMID: 37494115 Free PMC article. Review.
-
Short-range C-signaling restricts cheating behavior during Myxococcus xanthus development.mBio. 2024 Nov 13;15(11):e0244024. doi: 10.1128/mbio.02440-24. Epub 2024 Oct 18. mBio. 2024. PMID: 39422488 Free PMC article.
-
Kin recognition and outer membrane exchange (OME) in myxobacteria.Curr Opin Microbiol. 2020 Aug;56:81-88. doi: 10.1016/j.mib.2020.07.003. Epub 2020 Aug 20. Curr Opin Microbiol. 2020. PMID: 32828979 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
