Herpes Simplex Virus 1 Lytic Infection Blocks MicroRNA (miRNA) Biogenesis at the Stage of Nuclear Export of Pre-miRNAs
- PMID: 30755517
- PMCID: PMC6372804
- DOI: 10.1128/mBio.02856-18
Herpes Simplex Virus 1 Lytic Infection Blocks MicroRNA (miRNA) Biogenesis at the Stage of Nuclear Export of Pre-miRNAs
Abstract
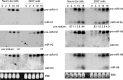
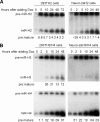
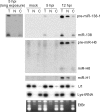
Herpes simplex virus 1 (HSV-1) switches between two infection programs, productive ("lytic") and latent infection. Some HSV-1 microRNAs (miRNAs) have been hypothesized to help control this switch, and yet little is known about regulation of their expression. Using Northern blot analyses, we found that, despite inherent differences in biogenesis efficiency among six HSV-1 miRNAs, all six exhibited high pre-miRNA/miRNA ratios during lytic infection of different cell lines and, when detectable, in acutely infected mouse trigeminal ganglia. In contrast, considerably lower ratios were observed in latently infected ganglia and in cells transduced with lentiviral vectors expressing the miRNAs, suggesting that HSV-1 lytic infection blocks miRNA biogenesis. This phenomenon is not specific to viral miRNAs, as a host miRNA expressed from recombinant HSV-1 also exhibited high pre-miRNA/miRNA ratios late during lytic infection. The levels of most of the mature miRNAs remained stable during infection in the presence of actinomycin D, indicating that the high ratios are due to inefficient pre-miRNA conversion to miRNA. Cellular fractionation experiments showed that late (but not early) during infection, pre-miRNAs were enriched in the nucleus and depleted in the cytoplasm, indicating that nuclear export was blocked. A mutation eliminating ICP27 expression or addition of acyclovir reduced pre-miRNA/miRNA ratios, but mutations drastically reducing Us11 expression did not. Thus, HSV-1 lytic infection inhibits miRNA biogenesis at the step of nuclear export and does so in an ICP27- and viral DNA synthesis-dependent manner. This mechanism may benefit the virus by reducing expression of repressive miRNAs during lytic infection while permitting elevated expression during latency.IMPORTANCE Various mechanisms have been identified by which viruses target host small RNA biogenesis pathways to achieve optimal infection outcomes. Herpes simplex virus 1 (HSV-1) is a ubiquitous human pathogen whose successful persistence in the host entails both productive ("lytic") and latent infection. Although many HSV-1 miRNAs have been discovered and some are thought to help control the lytic/latent switch, little is known about regulation of their biogenesis. By characterizing expression of both pre-miRNAs and mature miRNAs under various conditions, this study revealed striking differences in miRNA biogenesis between lytic and latent infection and uncovered a regulatory mechanism that blocks pre-miRNA nuclear export and is dependent on viral protein ICP27 and viral DNA synthesis. This mechanism represents a new virus-host interaction that could limit the repressive effects of HSV-1 miRNAs hypothesized to promote latency and may shed light on the regulation of miRNA nuclear export, which has been relatively unexplored.
Keywords: ICP27; herpes simplex virus; latency; miRNAs; nuclear export.
Copyright © 2019 Pan et al.
Figures




Similar articles
-
Mutations Inactivating Herpes Simplex Virus 1 MicroRNA miR-H2 Do Not Detectably Increase ICP0 Gene Expression in Infected Cultured Cells or Mouse Trigeminal Ganglia.J Virol. 2017 Jan 3;91(2):e02001-16. doi: 10.1128/JVI.02001-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27847363 Free PMC article.
-
A Herpesviral Lytic Protein Regulates the Structure of Latent Viral Chromatin.mBio. 2016 May 17;7(3):e00633-16. doi: 10.1128/mBio.00633-16. mBio. 2016. PMID: 27190217 Free PMC article.
-
Increased neurovirulence and reactivation of the herpes simplex virus type 1 latency-associated transcript (LAT)-negative mutant dLAT2903 with a disrupted LAT miR-H2.J Neurovirol. 2016 Feb;22(1):38-49. doi: 10.1007/s13365-015-0362-y. Epub 2015 Jun 12. J Neurovirol. 2016. PMID: 26069184 Free PMC article.
-
Control of HSV-1 latency in human trigeminal ganglia--current overview.J Neurovirol. 2011 Dec;17(6):518-27. doi: 10.1007/s13365-011-0063-0. Epub 2011 Dec 3. J Neurovirol. 2011. PMID: 22139603 Review.
-
Herpes Simplex Virus 1 MicroRNAs: An Update.Intervirology. 2023;66(1):97-110. doi: 10.1159/000531348. Epub 2023 Jun 7. Intervirology. 2023. PMID: 37285807 Free PMC article. Review.
Cited by
-
Deletion of Herpes Simplex Virus 1 MicroRNAs miR-H1 and miR-H6 Impairs Reactivation.J Virol. 2020 Jul 16;94(15):e00639-20. doi: 10.1128/JVI.00639-20. Print 2020 Jul 16. J Virol. 2020. PMID: 32295910 Free PMC article.
-
Neuronal miR-138 Represses HSV-2 Lytic Infection by Regulating Viral and Host Genes with Mechanistic Differences from HSV-1.J Virol. 2022 May 11;96(9):e0034922. doi: 10.1128/jvi.00349-22. Epub 2022 Apr 11. J Virol. 2022. PMID: 35404085 Free PMC article.
-
Interactome and Ubiquitinome Analyses Identify Functional Targets of Herpes Simplex Virus 1 Infected Cell Protein 0.Front Microbiol. 2022 Apr 18;13:856471. doi: 10.3389/fmicb.2022.856471. eCollection 2022. Front Microbiol. 2022. PMID: 35516420 Free PMC article.
-
Genes Involved in miRNA Biogenesis Are Not Downregulated in SARS-CoV-2 Infection.Viruses. 2023 May 16;15(5):1177. doi: 10.3390/v15051177. Viruses. 2023. PMID: 37243263 Free PMC article.
-
Rotavirus Induces Epithelial-Mesenchymal Transition Markers by Transcriptional Suppression of miRNA-29b.Front Microbiol. 2021 Feb 18;12:631183. doi: 10.3389/fmicb.2021.631183. eCollection 2021. Front Microbiol. 2021. PMID: 33679655 Free PMC article.
References
-
- Roizman B, Knipe DM, Whitley RJ. 2013. Herpes simplex virus, p 1823–1897. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racianello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
