Structural basis of ECF-σ-factor-dependent transcription initiation
- PMID: 30755604
- PMCID: PMC6372665
- DOI: 10.1038/s41467-019-08443-3
Structural basis of ECF-σ-factor-dependent transcription initiation
Abstract
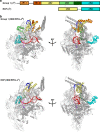
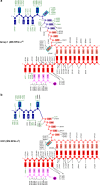
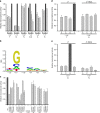
Extracytoplasmic (ECF) σ factors, the largest class of alternative σ factors, are related to primary σ factors, but have simpler structures, comprising only two of six conserved functional modules in primary σ factors: region 2 (σR2) and region 4 (σR4). Here, we report crystal structures of transcription initiation complexes containing Mycobacterium tuberculosis RNA polymerase (RNAP), M. tuberculosis ECF σ factor σL, and promoter DNA. The structures show that σR2 and σR4 of the ECF σ factor occupy the same sites on RNAP as in primary σ factors, show that the connector between σR2 and σR4 of the ECF σ factor-although shorter and unrelated in sequence-follows the same path through RNAP as in primary σ factors, and show that the ECF σ factor uses the same strategy to bind and unwind promoter DNA as primary σ factors. The results define protein-protein and protein-DNA interactions involved in ECF-σ-factor-dependent transcription initiation.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

