Biallelic VARS variants cause developmental encephalopathy with microcephaly that is recapitulated in vars knockout zebrafish
- PMID: 30755616
- PMCID: PMC6372652
- DOI: 10.1038/s41467-018-07953-w
Biallelic VARS variants cause developmental encephalopathy with microcephaly that is recapitulated in vars knockout zebrafish
Abstract
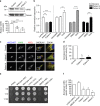
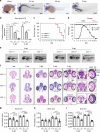
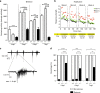
Aminoacyl tRNA synthetases (ARSs) link specific amino acids with their cognate transfer RNAs in a critical early step of protein translation. Mutations in ARSs have emerged as a cause of recessive, often complex neurological disease traits. Here we report an allelic series consisting of seven novel and two previously reported biallelic variants in valyl-tRNA synthetase (VARS) in ten patients with a developmental encephalopathy with microcephaly, often associated with early-onset epilepsy. In silico, in vitro, and yeast complementation assays demonstrate that the underlying pathomechanism of these mutations is most likely a loss of protein function. Zebrafish modeling accurately recapitulated some of the key neurological disease traits. These results provide both genetic and biological insights into neurodevelopmental disease and pave the way for further in-depth research on ARS related recessive disorders and precision therapies.
Conflict of interest statement
J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen, Inc., is on the Scientific Advisory Board of Baylor Genetic (BG) and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from molecular genetic testing offered in the BG Laboratories. All the remaining authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

