Screening of endogenous strong promoters for enhanced production of medium-chain-length polyhydroxyalkanoates in Pseudomonas mendocina NK-01
- PMID: 30755729
- PMCID: PMC6372614
- DOI: 10.1038/s41598-019-39321-z
Screening of endogenous strong promoters for enhanced production of medium-chain-length polyhydroxyalkanoates in Pseudomonas mendocina NK-01
Abstract
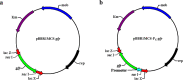
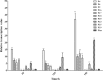
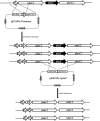
Polyhydroxyalkanoate (PHA) can be produced by microorganisms from renewable resources and is regarded as a promising bioplastic to replace petroleum-based plastics. Pseudomonas mendocina NK-01 is a medium-chain-length PHA (mcl-PHA)-producing strain and its whole-genome sequence is currently available. The yield of mcl-PHA in P. mendocina NK-01 is expected to be improved by applying a promoter engineering strategy. However, a limited number of well-characterized promoters has greatly restricted the application of promoter engineering for increasing the yield of mcl-PHA in P. mendocina NK-01. In this work, 10 endogenous promoters from P. mendocina NK-01 were identified based on RNA-seq and promoter prediction results. Subsequently, 10 putative promoters were characterized for their strength through the expression of a reporter gene gfp. As a result, five strong promoters designated as P4, P6, P9, P16 and P25 were identified based on transcriptional level and GFP fluorescence intensity measurements. To evaluate whether the screened promoters can be used to enhance transcription of PHA synthase gene (phaC), the three promoters P4, P6 and P16 were separately integrated into upstream of the phaC operon in the genome of P. mendocina NK-01, resulting in the recombinant strains NKU-4C1, NKU-6C1 and NKU-16C1. As expected, the transcriptional levels of phaC1 and phaC2 in the recombinant strains were increased as shown by real-time quantitative RT-PCR. The phaZ gene encoding PHA depolymerase was further deleted to construct the recombinant strains NKU-∆phaZ-4C1, NKU-∆phaZ-6C1 and NKU-∆phaZ-16C1. The results from shake-flask fermentation indicated that the mcl-PHA titer of recombinant strain NKU-∆phaZ-16C1 was increased from 17 to 23 wt% compared with strain NKU-∆phaZ. This work provides a feasible method to discover strong promoters in P. mendocina NK-01 and highlights the potential of the screened endogenous strong promoters for metabolic engineering of P. mendocina NK-01 to increase the yield of mcl-PHA.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Morphology engineering for enhanced production of medium-chain-length polyhydroxyalkanoates in Pseudomonas mendocina NK-01.Appl Microbiol Biotechnol. 2019 Feb;103(4):1713-1724. doi: 10.1007/s00253-018-9546-8. Epub 2019 Jan 4. Appl Microbiol Biotechnol. 2019. PMID: 30610286
-
Metabolic engineering of Pseudomonas mendocina NK-01 for enhanced production of medium-chain-length polyhydroxyalkanoates with enriched content of the dominant monomer.Int J Biol Macromol. 2020 Jul 1;154:1596-1605. doi: 10.1016/j.ijbiomac.2019.11.044. Epub 2019 Nov 7. Int J Biol Macromol. 2020. PMID: 31706817
-
A promoter engineering-based strategy enhances polyhydroxyalkanoate production in Pseudomonas putida KT2440.Int J Biol Macromol. 2021 Nov 30;191:608-617. doi: 10.1016/j.ijbiomac.2021.09.142. Epub 2021 Sep 25. Int J Biol Macromol. 2021. PMID: 34582907
-
PHA synthase (PhaC): interpreting the functions of bioplastic-producing enzyme from a structural perspective.Appl Microbiol Biotechnol. 2019 Feb;103(3):1131-1141. doi: 10.1007/s00253-018-9538-8. Epub 2018 Dec 3. Appl Microbiol Biotechnol. 2019. PMID: 30511262 Review.
-
A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry.Chem Soc Rev. 2009 Aug;38(8):2434-46. doi: 10.1039/b812677c. Epub 2009 May 8. Chem Soc Rev. 2009. PMID: 19623359 Review.
Cited by
-
In silico identification of bacterial seaweed-degrading bioplastic producers.Microb Genom. 2022 Sep;8(9):mgen000866. doi: 10.1099/mgen.0.000866. Microb Genom. 2022. PMID: 36125959 Free PMC article.
-
A review on enzymes and pathways for manufacturing polyhydroxybutyrate from lignocellulosic materials.3 Biotech. 2021 Nov;11(11):483. doi: 10.1007/s13205-021-03009-x. Epub 2021 Oct 30. 3 Biotech. 2021. PMID: 34790507 Free PMC article. Review.
-
Advancements in genetic engineering for enhanced Polyhydroxyalkanoates (PHA) production: a comprehensive review of metabolic pathway manipulation and gene deletion strategies.Bioengineered. 2025 Dec;16(1):2458363. doi: 10.1080/21655979.2025.2458363. Epub 2025 Jan 30. Bioengineered. 2025. PMID: 39882623 Free PMC article. Review.
-
Advances in Microbial Biotechnology for Sustainable Alternatives to Petroleum-Based Plastics: A Comprehensive Review of Polyhydroxyalkanoate Production.Microorganisms. 2024 Aug 13;12(8):1668. doi: 10.3390/microorganisms12081668. Microorganisms. 2024. PMID: 39203509 Free PMC article. Review.
-
Exploring the Role of Green Microbes in Sustainable Bioproduction of Biodegradable Polymers.Polymers (Basel). 2023 Dec 4;15(23):4617. doi: 10.3390/polym15234617. Polymers (Basel). 2023. PMID: 38232039 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

