A Top-Down Proteomics Platform Coupling Serial Size Exclusion Chromatography and Fourier Transform Ion Cyclotron Resonance Mass Spectrometry
- PMID: 30758949
- PMCID: PMC6545233
- DOI: 10.1021/acs.analchem.8b04082
A Top-Down Proteomics Platform Coupling Serial Size Exclusion Chromatography and Fourier Transform Ion Cyclotron Resonance Mass Spectrometry
Abstract
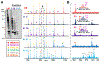
Mass spectrometry (MS) based top-down proteomics provides rich information about proteoforms arising from combinatorial amino acid sequence variations and post-translational modifications (PTMs). Fourier transform ion cyclotron resonance (FT-ICR) MS affords ultrahigh resolving power and provides high-accuracy mass measurements, presenting a powerful tool for top-down MS characterization of proteoforms. However, the detection and characterization of large proteins from complex mixtures remain challenging due to the exponential decrease in S: N with increasing molecular weight (MW) and coeluting low-MW proteins; thus, size-based fractionation of complex protein mixtures prior to MS analysis is necessary. Here, we directly combine MS-compatible serial size exclusion chromatography (sSEC) fractionation with 12 T FT-ICR MS for targeted top-down characterization of proteins from complex mixtures extracted from human and swine heart tissue. Benefiting from the ultrahigh resolving power of FT-ICR, we isotopically resolved 31 distinct proteoforms (30-50 kDa) simultaneously in a single mass spectrum within a 100 m/ z window. Notably, within a 5 m/ z window, we obtained baseline isotopic resolution for 6 distinct large proteoforms (30-50 kDa). The ultrahigh resolving power of FT-ICR MS combined with sSEC fractionation enabled targeted top-down analysis of large proteoforms (>30 kDa) from the human heart proteome without extensive chromatographic separation or protein purification. Further separation of proteoforms inside the mass spectrometer (in-MS) allowed for isolation of individual proteoforms and targeted electron capture dissociation (ECD), yielding high sequence coverage. sSEC/FT-ICR ECD facilitated the identification and sequence characterization of important metabolic enzymes. This platform, which facilitates deep interrogation of proteoform primary structure, is highly tunable, allows for adjustment of MS and MS/MS parameters in real time, and can be utilized for a variety of complex protein mixtures.
Figures



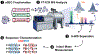
Similar articles
-
Fourier-transform ion cyclotron resonance mass spectrometry for characterizing proteoforms.Mass Spectrom Rev. 2022 Mar;41(2):158-177. doi: 10.1002/mas.21653. Epub 2020 Sep 7. Mass Spectrom Rev. 2022. PMID: 32894796 Free PMC article. Review.
-
Top-Down Proteomics of Large Proteins up to 223 kDa Enabled by Serial Size Exclusion Chromatography Strategy.Anal Chem. 2017 May 16;89(10):5467-5475. doi: 10.1021/acs.analchem.7b00380. Epub 2017 May 2. Anal Chem. 2017. PMID: 28406609 Free PMC article.
-
Identification and Characterization of Human Proteoforms by Top-Down LC-21 Tesla FT-ICR Mass Spectrometry.J Proteome Res. 2017 Feb 3;16(2):1087-1096. doi: 10.1021/acs.jproteome.6b00696. Epub 2016 Dec 12. J Proteome Res. 2017. PMID: 27936753 Free PMC article.
-
Intact-Mass Analysis Facilitating the Identification of Large Human Heart Proteoforms.Anal Chem. 2019 Sep 3;91(17):10937-10942. doi: 10.1021/acs.analchem.9b02343. Epub 2019 Aug 14. Anal Chem. 2019. PMID: 31393705 Free PMC article.
-
Towards analytically useful two-dimensional Fourier transform ion cyclotron resonance mass spectrometry.Anal Bioanal Chem. 2013 Jan;405(1):51-61. doi: 10.1007/s00216-012-6422-8. Epub 2012 Oct 18. Anal Bioanal Chem. 2013. PMID: 23076397 Review.
Cited by
-
Native top-down proteomics enables discovery in endocrine-resistant breast cancer.Nat Chem Biol. 2025 Aug;21(8):1205-1213. doi: 10.1038/s41589-025-01866-8. Epub 2025 Apr 4. Nat Chem Biol. 2025. PMID: 40186031
-
Capillary Zone Electrophoresis-Electron-Capture Collision-Induced Dissociation on a Quadrupole Time-of-Flight Mass Spectrometer for Top-Down Characterization of Intact Proteins.J Am Soc Mass Spectrom. 2021 Jun 2;32(6):1361-1369. doi: 10.1021/jasms.0c00484. Epub 2021 Mar 22. J Am Soc Mass Spectrom. 2021. PMID: 33749270 Free PMC article.
-
Fourier-transform ion cyclotron resonance mass spectrometry for characterizing proteoforms.Mass Spectrom Rev. 2022 Mar;41(2):158-177. doi: 10.1002/mas.21653. Epub 2020 Sep 7. Mass Spectrom Rev. 2022. PMID: 32894796 Free PMC article. Review.
-
Capillary Zone Electrophoresis-Tandem Mass Spectrometry for Top-Down Proteomics of Mouse Brain Integral Membrane Proteins.Anal Chem. 2023 Aug 29;95(34):12590-12594. doi: 10.1021/acs.analchem.3c02346. Epub 2023 Aug 18. Anal Chem. 2023. PMID: 37595263 Free PMC article.
-
High-throughput quantitative top-down proteomics.Mol Omics. 2020 Apr 1;16(2):91-99. doi: 10.1039/c9mo00154a. Epub 2020 Jan 14. Mol Omics. 2020. PMID: 31932818 Free PMC article. Review.
References
-
- Aebersold R; Agar JN; Amster IJ; Baker MS; Bertozzi CR; Boja ES; Costello CE; Cravatt BF; Fenselau C; Garcia BA; Ge Y; Gunawardena J; Hendrickson RC; Hergenrother PJ; Huber CG; Ivanov AR; Jensen ON; Jewett MC; Kelleher NL; Kiessling LL; Krogan NJ; Larsen MR; Loo JA; Ogorzalek Loo RR; Lundberg E; MacCoss MJ; Mallick P; Mootha VK; Mrksich M; Muir TW; Patrie SM; Pesavento JJ; Pitteri SJ; Rodriguez H; Saghatelian A; Sandoval W; Schluter H; Sechi S; Slavoff SA; Smith LM; Snyder MP; Thomas PM; Uhlen M; Van Eyk JE; Vidal M; Walt DR; White FM; Williams ER; Wohlschlager T; Wysocki VH; Yates NA; Young NL; Zhang B, How many human proteoforms are there? Nat Chem Biol 2018, 14 (3), 206–214. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

