Structural organization of a Type III-A CRISPR effector subcomplex determined by X-ray crystallography and cryo-EM
- PMID: 30759237
- PMCID: PMC6468305
- DOI: 10.1093/nar/gkz079
Structural organization of a Type III-A CRISPR effector subcomplex determined by X-ray crystallography and cryo-EM
Abstract
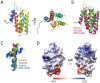
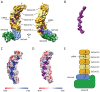
Clustered regularly interspaced short palindromic repeats (CRISPR) and their associated Cas proteins provide an immune-like response in many prokaryotes against extraneous nucleic acids. CRISPR-Cas systems are classified into different classes and types. Class 1 CRISPR-Cas systems form multi-protein effector complexes that includes a guide RNA (crRNA) used to identify the target for destruction. Here we present crystal structures of Staphylococcus epidermidis Type III-A CRISPR subunits Csm2 and Csm3 and a 5.2 Å resolution single-particle cryo-electron microscopy (cryo-EM) reconstruction of an in vivo assembled effector subcomplex including the crRNA. The structures help to clarify the quaternary architecture of Type III-A effector complexes, and provide details on crRNA binding, target RNA binding and cleavage, and intermolecular interactions essential for effector complex assembly. The structures allow a better understanding of the organization of Type III-A CRISPR effector complexes as well as highlighting the overall similarities and differences with other Class 1 effector complexes.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Structures of an active type III-A CRISPR effector complex.Structure. 2022 Aug 4;30(8):1109-1128.e6. doi: 10.1016/j.str.2022.05.013. Epub 2022 Jun 16. Structure. 2022. PMID: 35714601 Free PMC article.
-
Crystal Structures of Csm2 and Csm3 in the Type III-A CRISPR-Cas Effector Complex.J Mol Biol. 2019 Feb 15;431(4):748-763. doi: 10.1016/j.jmb.2019.01.009. Epub 2019 Jan 11. J Mol Biol. 2019. PMID: 30639408
-
Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.Mol Cell. 2019 Jan 17;73(2):264-277.e5. doi: 10.1016/j.molcel.2018.11.007. Epub 2018 Nov 29. Mol Cell. 2019. PMID: 30503773 Free PMC article.
-
Molecular Mechanisms of RNA Targeting by Cas13-containing Type VI CRISPR-Cas Systems.J Mol Biol. 2019 Jan 4;431(1):66-87. doi: 10.1016/j.jmb.2018.06.029. Epub 2018 Jun 22. J Mol Biol. 2019. PMID: 29940185 Review.
-
Approaches to study CRISPR RNA biogenesis and the key players involved.Methods. 2020 Feb 1;172:12-26. doi: 10.1016/j.ymeth.2019.07.015. Epub 2019 Jul 17. Methods. 2020. PMID: 31325492 Review.
Cited by
-
Critical roles for 'housekeeping' nucleases in type III CRISPR-Cas immunity.Elife. 2022 Dec 8;11:e81897. doi: 10.7554/eLife.81897. Elife. 2022. PMID: 36479971 Free PMC article.
-
CRISPR/dCas9-Based Systems: Mechanisms and Applications in Plant Sciences.Plants (Basel). 2021 Sep 29;10(10):2055. doi: 10.3390/plants10102055. Plants (Basel). 2021. PMID: 34685863 Free PMC article. Review.
-
Cas10 relieves host growth arrest to facilitate spacer retention during type III-A CRISPR-Cas immunity.Cell Host Microbe. 2024 Dec 11;32(12):2050-2062.e6. doi: 10.1016/j.chom.2024.11.005. Epub 2024 Dec 2. Cell Host Microbe. 2024. PMID: 39626678 Free PMC article.
-
Type III-A CRISPR systems as a versatile gene knockdown technology.RNA. 2022 Aug;28(8):1074-1088. doi: 10.1261/rna.079206.122. Epub 2022 May 26. RNA. 2022. PMID: 35618430 Free PMC article.
-
Structures of an active type III-A CRISPR effector complex.Structure. 2022 Aug 4;30(8):1109-1128.e6. doi: 10.1016/j.str.2022.05.013. Epub 2022 Jun 16. Structure. 2022. PMID: 35714601 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

