Temporal proteomic profiling reveals changes that support Burkholderia biofilms
- PMID: 30759239
- PMCID: PMC6482045
- DOI: 10.1093/femspd/ftz005
Temporal proteomic profiling reveals changes that support Burkholderia biofilms
Abstract
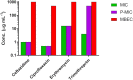
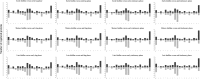
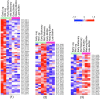
Melioidosis associated with opportunistic pathogen Burkholderia pseudomallei imparts a huge medical burden in Southeast Asia and Australia. At present there is no available human vaccine that protects against B. pseudomallei infection and antibiotic treatments are limited particularly for drug-resistant strains and bacteria in biofilm forms. Biofilm forming bacteria exhibit phenotypic features drastically different to their planktonic states, often exhibiting a diminished response to antimicrobial therapies. Our earlier work on global profiling of bacterial biofilms using transcriptomics and proteomics revealed transcript-decoupled protein abundance in bacterial biofilms. Here we employed reverse phase liquid chromatography tandem mass spectrometry (LC-MS/MS) to deduce temporal proteomic differences in planktonic and biofilm forms of Burkholderia thailandensis, which is weakly surrogate model of pathogenic B. pseudomallei as sharing a key element in genomic similarity. The proteomic analysis of B. thailandensis in biofilm versus planktonic states revealed that proteome changes support biofilm survival through decreased abundance of metabolic proteins while increased abundance of stress-related proteins. Interestingly, the protein abundance including for the transcription protein TEX, outer periplasmic TolB protein, and the exopolyphosphatase reveal adaption in bacterial biofilms that facilitate antibiotic tolerance through a non-specific mechanism. The present proteomics study of B. thailandensis biofilms provides a global snapshot of protein abundance differences and antimicrobial sensitivities in planktonic and sessile bacteria.
Keywords: antimicrobial tolerance; biofilms; liquid chromatography mass spectrometry; melioidosis; proteomics.
Published by Oxford University Press on behalf of FEMS 2019.
Figures


References
-
- Adrien T, Thorsten M, Thomas B et al. Membrane chaperoning by members of the PspA/IM30 protein family. Commun Integr Biol. 2017;10:e1264546.

