Diurnal Rhythms Spatially and Temporally Organize Autophagy
- PMID: 30759397
- PMCID: PMC6442472
- DOI: 10.1016/j.celrep.2019.01.072
Diurnal Rhythms Spatially and Temporally Organize Autophagy
Abstract
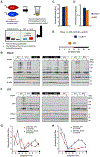
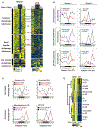
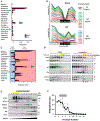
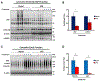
Circadian rhythms are a hallmark of physiology, but how such daily rhythms organize cellular catabolism is poorly understood. Here, we used proteomics to map daily oscillations in autophagic flux in mouse liver and related these rhythms to proteasome activity. We also explored how systemic inflammation affects the temporal structure of autophagy. Our data identified a globally harmonized rhythm for basal macroautophagy, chaperone-mediated autophagy, and proteasomal activity, which concentrates liver proteolysis during the daytime. Basal autophagy rhythms could be resolved into two antiphase clusters that were distinguished by the subcellular location of targeted proteins. Inflammation induced by lipopolysaccharide reprogrammed autophagic flux away from a temporal pattern that favors cytosolic targets and toward the turnover of mitochondrial targets. Our data detail how daily biological rhythms connect the temporal, spatial, and metabolic aspects of protein catabolism.
Keywords: autophagy; chaperone-mediated autophagy; circadian rhythm; clock; endotoxin; inflammation; lipopolysaccharide; macroautophagy; proteasome.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
A.M.K.C. is a cofounder, stock holder, and serves on the Scientific Advisory Board for Proterris, which develops therapeutic uses for carbon monoxide. A.M.K.C. also has a use patent on CO. A.M.K.C. served as a consultant for Teva Pharmaceuticals in July 2018.
Figures


Comment in
-
Adventures in spacetime: circadian rhythms and the dynamics of protein catabolism.Autophagy. 2019 Jun;15(6):1115-1116. doi: 10.1080/15548627.2019.1596498. Epub 2019 Mar 27. Autophagy. 2019. PMID: 30894057 Free PMC article.
References
-
- Agarraberes FA, and Dice JF (2001). A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J. Cell Sci 774, 2491–2499. - PubMed
-
- Asano S, Fukuda Y, Beck F, Aufderheide A, Förster F, Danev R, and Baumeister W (2015). Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science 347, 439–442. - PubMed
-
- Chittum HS, Lane WS, Carlson BA, Roller PP, Lung FD, Lee BJ, and Hatfield DL (1998). Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry 37, 10866–10870. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

