Light-Induced Protein Clustering for Optogenetic Interference and Protein Interaction Analysis in Drosophila S2 Cells
- PMID: 30759894
- PMCID: PMC6406598
- DOI: 10.3390/biom9020061
Light-Induced Protein Clustering for Optogenetic Interference and Protein Interaction Analysis in Drosophila S2 Cells
Abstract
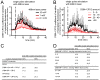
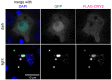
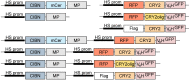
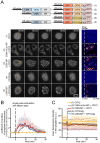
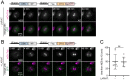
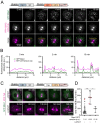
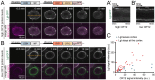
Drosophila Schneider 2 (S2) cells are a simple and powerful system commonly used in cell biology because they are well suited for high resolution microscopy and RNAi-mediated depletion. However, understanding dynamic processes, such as cell division, also requires methodology to interfere with protein function with high spatiotemporal control. In this research study, we report the adaptation of an optogenetic tool to Drosophila S2 cells. Light-activated reversible inhibition by assembled trap (LARIAT) relies on the rapid light-dependent heterodimerization between cryptochrome 2 (CRY2) and cryptochrome-interacting bHLH 1 (CIB1) to form large protein clusters. An anti-green fluorescent protein (GFP) nanobody fused with CRY2 allows this method to quickly trap any GFP-tagged protein in these light-induced protein clusters. We evaluated clustering kinetics in response to light for different LARIAT modules, and showed the ability of GFP-LARIAT to inactivate the mitotic protein Mps1 and to disrupt the membrane localization of the polarity regulator Lethal Giant Larvae (Lgl). Moreover, we validated light-induced co-clustering assays to assess protein-protein interactions in S2 cells. In conclusion, GFP-based LARIAT is a versatile tool to answer different biological questions, since it enables probing of dynamic processes and protein-protein interactions with high spatiotemporal resolution in Drosophila S2 cells.
Keywords: Drosophila; LARIAT; Mps1; Schneider 2 cells; aPKC; cell polarity; mitosis; optogenetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Protein Inactivation by Optogenetic Trapping in Living Cells.Methods Mol Biol. 2016;1408:363-76. doi: 10.1007/978-1-4939-3512-3_25. Methods Mol Biol. 2016. PMID: 26965136
-
The Dual Characteristics of Light-Induced Cryptochrome 2, Homo-oligomerization and Heterodimerization, for Optogenetic Manipulation in Mammalian Cells.ACS Synth Biol. 2015 Oct 16;4(10):1124-35. doi: 10.1021/acssynbio.5b00048. Epub 2015 Jun 8. ACS Synth Biol. 2015. PMID: 25985220 Free PMC article.
-
Red Light-Activated Reversible Inhibition of Protein Functions by Assembled Trap.ACS Synth Biol. 2025 May 16;14(5):1437-1450. doi: 10.1021/acssynbio.4c00585. Epub 2025 Apr 30. ACS Synth Biol. 2025. PMID: 40304578
-
deGradFP: A System to Knockdown GFP-Tagged Proteins.Methods Mol Biol. 2016;1478:177-187. doi: 10.1007/978-1-4939-6371-3_9. Methods Mol Biol. 2016. PMID: 27730581 Review.
-
Connecting epithelial polarity, proliferation and cancer in Drosophila: the many faces of lgl loss of function.Int J Dev Biol. 2013;57(9-10):677-87. doi: 10.1387/ijdb.130285dg. Int J Dev Biol. 2013. PMID: 24395559 Review.
Cited by
-
Recent evolution of a TET-controlled and DPPA3/STELLA-driven pathway of passive DNA demethylation in mammals.Nat Commun. 2020 Nov 24;11(1):5972. doi: 10.1038/s41467-020-19603-1. Nat Commun. 2020. PMID: 33235224 Free PMC article.
-
Nanobody-Based Probes for Subcellular Protein Identification and Visualization.Front Cell Neurosci. 2020 Nov 2;14:573278. doi: 10.3389/fncel.2020.573278. eCollection 2020. Front Cell Neurosci. 2020. PMID: 33240044 Free PMC article. Review.
-
A light-inducible protein clustering system for in vivo analysis of α-synuclein aggregation in Parkinson disease.PLoS Biol. 2022 Mar 9;20(3):e3001578. doi: 10.1371/journal.pbio.3001578. eCollection 2022 Mar. PLoS Biol. 2022. PMID: 35263320 Free PMC article.
-
Mps1-mediated release of Mad1 from nuclear pores ensures the fidelity of chromosome segregation.J Cell Biol. 2020 Mar 2;219(3):e201906039. doi: 10.1083/jcb.201906039. J Cell Biol. 2020. PMID: 31913420 Free PMC article.
-
CeLINC, a fluorescence-based protein-protein interaction assay in Caenorhabditis elegans.Genetics. 2021 Dec 10;219(4):iyab163. doi: 10.1093/genetics/iyab163. Genetics. 2021. PMID: 34849800 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

