Identification and characterization of genes with absolute mRNA abundances changes in tumor cells with varied transcriptome sizes
- PMID: 30760197
- PMCID: PMC6374894
- DOI: 10.1186/s12864-019-5502-y
Identification and characterization of genes with absolute mRNA abundances changes in tumor cells with varied transcriptome sizes
Abstract
Background: The amount of RNA per cell, namely the transcriptome size, may vary under many biological conditions including tumor. If the transcriptome size of two cells is different, direct comparison of the expression measurements on the same amount of total RNA for two samples can only identify genes with changes in the relative mRNA abundances, i.e., cellular mRNA concentration, rather than genes with changes in the absolute mRNA abundances.
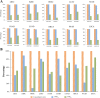
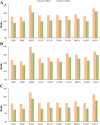
Results: Our recently proposed RankCompV2 algorithm identify differentially expressed genes (DEGs) through comparing the relative expression orderings (REOs) of disease samples with that of normal samples. We reasoned that both the mRNA concentration and the absolute abundances of these DEGs must have changes in disease samples. In simulation experiments, this method showed excellent performance for identifying DEGs between normal and disease samples with different transcriptome sizes. Through analyzing data for ten cancer types, we found that a significantly higher proportion of the DEGs with absolute mRNA abundance changes overlapped or directly interacted with known cancer driver genes and anti-cancer drug targets than that of the DEGs only with mRNA concentration changes alone identified by the traditional methods. The DEGs with increased absolute mRNA abundances were enriched in DNA damage-related pathways, while DEGs with decreased absolute mRNA abundances were enriched in immune and metabolism associated pathways.
Conclusions: Both the mRNA concentration and the absolute abundances of the DEGs identified through REOs comparison change in disease samples in comparison with normal samples. In cancers these genes might play more important upstream roles in carcinogenesis.
Keywords: Absolute mRNA abundances; Cellular mRNA concentration; Differentially expressed gene; Relative expression ordering.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Birchler JA. Facts and artifacts in studies of gene expression in aneuploids and sex chromosomes. Chromosoma. 2014;123(5):459–469. - PubMed
-
- Stevens JB, Horne SD, Abdallah BY, Ye CJ, Heng HH. Chromosomal instability and transcriptome dynamics in cancer. Cancer Metastasis Rev. 2013;32(3–4):391–402. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

