Adaptation of an R5 Simian-Human Immunodeficiency Virus Encoding an HIV Clade A Envelope with or without Ablation of Adaptive Host Immunity: Differential Selection of Viral Mutants
- PMID: 30760566
- PMCID: PMC6475780
- DOI: 10.1128/JVI.02267-18
Adaptation of an R5 Simian-Human Immunodeficiency Virus Encoding an HIV Clade A Envelope with or without Ablation of Adaptive Host Immunity: Differential Selection of Viral Mutants
Abstract
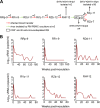
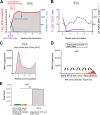
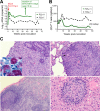
Simian-human immunodeficiency virus (SHIV) infection in rhesus macaques (RMs) resembles human immunodeficiency virus type 1 (HIV-1) infection in humans and serves as a tool to evaluate candidate AIDS vaccines. HIV-1 clade A (HIV-A) predominates in parts of Africa. We constructed an R5 clade A SHIV (SHIV-A; strain SHIV-KNH1144) carrying env from a Kenyan HIV-A. SHIV-A underwent rapid serial passage through six RMs. To allow unbridled replication without adaptive immunity, we simultaneously ablated CD8+ and B cells with cytotoxic monoclonal antibodies in the next RM, resulting in extremely high viremia and CD4+ T-cell loss. Infected blood was then transferred into two non-immune-depleted RMs, where progeny SHIV-A showed increased replicative capacity and caused AIDS. We reisolated SHIV-KNH1144p4, which was replication competent in peripheral blood mononuclear cells (PBMC) of all RMs tested. Next-generation sequencing of early- and late-passage SHIV-A strains identified mutations that arose due to "fitness" virus optimization in the former and mutations exhibiting signatures typical for adaptive host immunity in the latter. "Fitness" mutations are best described as mutations that allow for better fit of the HIV-A Env with SIV-derived virion building blocks or host proteins and mutations in noncoding regions that accelerate virus replication, all of which result in the outgrowth of virus variants in the absence of adaptive T-cell and antibody-mediated host immunity.IMPORTANCE In this study, we constructed a simian-human immunodeficiency virus carrying an R5 Kenyan HIV-1 clade A env (SHIV-A). To bypass host immunity, SHIV-A was rapidly passaged in naive macaques or animals depleted of both CD8+ and B cells. Next-generation sequencing identified different mutations that resulted from optimization of viral replicative fitness either in the absence of adaptive immunity or due to pressure from adaptive immune responses.
Keywords: HIV; SHIV; SHIV-A; adaptation; immunodepletion; rhesus macaques.
Copyright © 2019 American Society for Microbiology.
Figures



References
-
- Hraber P, Korber BT, Lapedes AS, Bailer RT, Seaman MS, Gao H, Greene KM, McCutchan F, Williamson C, Kim JH, Tovanabutra S, Hahn BH, Swanstrom R, Thomson MM, Gao F, Harris L, Giorgi E, Hengartner N, Bhattacharya T, Mascola JR, Montefiori DC. 2014. Impact of clade, geography, and age of the epidemic on HIV-1 neutralization by antibodies. J Virol 88:12623–12643. doi:10.1128/JVI.01705-14. - DOI - PMC - PubMed
-
- Rainwater S, DeVange S, Sagar M, Ndinya-Achola J, Mandaliya K, Kreiss JK, Overbaugh J. 2005. No evidence for rapid subtype C spread within an epidemic in which multiple subtypes and intersubtype recombinants circulate. AIDS Res Hum Retroviruses 21:1060–1065. doi:10.1089/aid.2005.21.1060. - DOI - PubMed
-
- Kiwanuka N, Laeyendecker O, Quinn TC, Wawer MJ, Shepherd J, Robb M, Kigozi G, Kagaayi J, Serwadda D, Makumbi FE, Reynolds SJ, Gray RH. 2009. HIV-1 subtypes and differences in heterosexual HIV transmission among HIV-discordant couples in Rakai, Uganda. AIDS 23:2479–2484. doi:10.1097/QAD.0b013e328330cc08. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

