Dose-Dependent Differences in HIV Inhibition by Different Interferon Alpha Subtypes While Having Overall Similar Biologic Effects
- PMID: 30760614
- PMCID: PMC6374594
- DOI: 10.1128/mSphere.00637-18
Dose-Dependent Differences in HIV Inhibition by Different Interferon Alpha Subtypes While Having Overall Similar Biologic Effects
Abstract
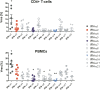
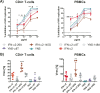
Type I interferons (IFNs) are key players in the antiviral immune response. Interferon alpha (IFN-α) belongs to this class of IFNs and comprises 12 subtypes that differ from each other in their binding affinities for a common receptor and, thus, in their signaling potencies. Recent data suggest that IFN-α6 and -α14 are the most potent IFN-α subtypes in restricting HIV replication when applied exogenously. However, in the context of antiviral therapy, IFNs are administered at high doses, which may compensate for differences in potency seen between IFN-α subtypes. In this study, we reexamined whether IFN-α subtypes induce different biological activities, with a focus on how IFN-α treatment dose affects cellular responses to HIV in primary CD4+ T cells, peripheral blood mononuclear cells (PBMCs), and macrophages. We found that the subtypes' antiviral activities were dose dependent, with >90% inhibition of HIV replication at a high dose of all IFN-αs except the weak IFN-α/β receptor (IFNAR) binder, IFN-α1. The quality of the responses engendered by IFN-α1, -α2, -α6, and -α14 was highly comparable, with essentially the same set of genes induced by all four subtypes. Hierarchal cluster analysis revealed that the individual donors were stronger determinants for the IFN-stimulated-gene (ISG) responses than the specific IFN-α subtype used for stimulation. Notably, IFN-α2-derived mutants with substantially reduced IFNAR2 binding still inhibited HIV replication efficiently, whereas mutants with increased IFNAR1 binding potentiated antiviral activity. Overall, our results support the idea that IFN-α subtypes do not induce different biological responses, given that each subtype is exogenously applied at bioequivalent doses.IMPORTANCE Elucidating the functional role of the IFN-α subtypes is of particular importance for the development of efficacious therapies using exogenous IFN-α. Specifically, this will help define whether IFN therapy should be based on the use of pathogen-dependent IFN subtypes or, rather, IFN mutants with optimized IFNAR binding properties.
Keywords: antiviral therapy; human immunodeficiency virus; interferons; therapeutic efficacy.
Copyright © 2019 Schlaepfer et al.
Figures


Comment in
-
Different Biological Activities of Specific Interferon Alpha Subtypes.mSphere. 2019 Jul 24;4(4):e00127-19. doi: 10.1128/mSphere.00127-19. mSphere. 2019. PMID: 31341068 Free PMC article. No abstract available.
References
-
- Sun H, Buzon MJ, Shaw A, Berg RK, Yu XG, Ferrando-Martinez S, Leal M, Ruiz-Mateos E, Lichterfeld M. 2014. Hepatitis C therapy with interferon-alpha and ribavirin reduces CD4 T-cell-associated HIV-1 DNA in HIV-1/hepatitis C virus-coinfected patients. J Infect Dis 209:1315–1320. doi:10.1093/infdis/jit628. - DOI - PMC - PubMed
-
- Azzoni L, Foulkes AS, Papasavvas E, Mexas AM, Lynn KM, Mounzer K, Tebas P, Jacobson JM, Frank I, Busch MP, Deeks SG, Carrington M, O'Doherty U, Kostman J, Montaner LJ. 2013. Pegylated interferon alfa-2a monotherapy results in suppression of HIV type 1 replication and decreased cell-associated HIV DNA integration. J Infect Dis 207:213–222. doi:10.1093/infdis/jis663. - DOI - PMC - PubMed
-
- Moron-Lopez S, Gomez-Mora E, Salgado M, Ouchi D, Puertas MC, Urrea V, Navarro J, Jou A, Perez M, Tural C, Clotet B, Montaner LJ, Blanco J, Crespo M, Martinez-Picado J. 2016. Short-term treatment with interferon alfa diminishes expression of HIV-1 and reduces CD4+ T-cell activation in patients coinfected with HIV and hepatitis C virus and receiving antiretroviral therapy. J Infect Dis 213:1008–1012. doi:10.1093/infdis/jiv521. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
