Basic Units of Inter-Individual Variation in Resting State Connectomes
- PMID: 30760808
- PMCID: PMC6374507
- DOI: 10.1038/s41598-018-38406-5
Basic Units of Inter-Individual Variation in Resting State Connectomes
Abstract
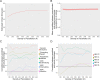
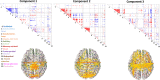
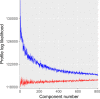
Resting state functional connectomes are massive and complex. It is an open question, however, whether connectomes differ across individuals in a correspondingly massive number of ways, or whether most differences take a small number of characteristic forms. We systematically investigated this question and found clear evidence of low-rank structure in which a modest number of connectomic components, around 50-150, account for a sizable portion of inter-individual connectomic variation. This number was convergently arrived at with multiple methods including estimation of intrinsic dimensionality and assessment of reconstruction of out-of-sample data. In addition, we show that these connectomic components enable prediction of a broad array of neurocognitive and clinical symptom variables at levels comparable to a leading method that is trained on the whole connectome. Qualitative observation reveals that these connectomic components exhibit extensive community structure reflecting interrelationships between intrinsic connectivity networks. We provide quantitative validation of this observation using novel stochastic block model-based methods. We propose that these connectivity components form an effective basis set for quantifying and interpreting inter-individual connectomic differences, and for predicting behavioral/clinical phenotypes.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

