Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death
- PMID: 30760928
- PMCID: PMC6405297
- DOI: 10.1038/s41586-019-0945-5
Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death
Abstract
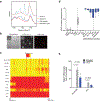
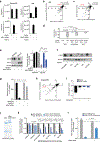
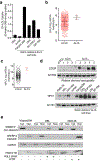
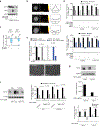
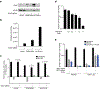
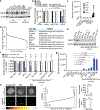
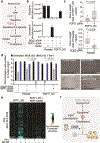
Cholesterol is essential for cells to grow and proliferate. Normal mammalian cells meet their need for cholesterol through its uptake or de novo synthesis1, but the extent to which cancer cells rely on each of these pathways remains poorly understood. Here, using a competitive proliferation assay on a pooled collection of DNA-barcoded cell lines, we identify a subset of cancer cells that is auxotrophic for cholesterol and thus highly dependent on its uptake. Through metabolic gene expression analysis, we pinpoint the loss of squalene monooxygenase expression as a cause of cholesterol auxotrophy, particularly in ALK+ anaplastic large cell lymphoma (ALCL) cell lines and primary tumours. Squalene monooxygenase catalyses the oxidation of squalene to 2,3-oxidosqualene in the cholesterol synthesis pathway and its loss results in accumulation of the upstream metabolite squalene, which is normally undetectable. In ALK+ ALCLs, squalene alters the cellular lipid profile and protects cancer cells from ferroptotic cell death, providing a growth advantage under conditions of oxidative stress and in tumour xenografts. Finally, a CRISPR-based genetic screen identified cholesterol uptake by the low-density lipoprotein receptor as essential for the growth of ALCL cells in culture and as patient-derived xenografts. This work reveals that the cholesterol auxotrophy of ALCLs is a targetable liability and, more broadly, that systematic approaches can be used to identify nutrient dependencies unique to individual cancer types.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures







References
-
- Kidd JG Regression of transplanted lymphomas induced in vivo by means of normal guinea pig serum. II. Studies on the nature of the active serum constituent: histological mechanism of the regression: tests for effects of guinea pig serum on lymphoma cells in vitro: discussion. J Exp Med 98, 583–606 (1953). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

