Exploring K2G30 Genome: A High Bacterial Cellulose Producing Strain in Glucose and Mannitol Based Media
- PMID: 30761107
- PMCID: PMC6363697
- DOI: 10.3389/fmicb.2019.00058
Exploring K2G30 Genome: A High Bacterial Cellulose Producing Strain in Glucose and Mannitol Based Media
Abstract
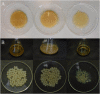
Demands for renewable and sustainable biopolymers have rapidly increased in the last decades along with environmental issues. In this context, bacterial cellulose, as renewable and biodegradable biopolymer has received considerable attention. Particularly, acetic acid bacteria of the Komagataeibacter xylinus species can produce bacterial cellulose from several carbon sources. To fully exploit metabolic potential of cellulose producing acetic acid bacteria, an understanding of the ability of producing bacterial cellulose from different carbon sources and the characterization of the genes involved in the synthesis is required. Here, K2G30 (UMCC 2756) was studied with respect to bacterial cellulose production in mannitol, xylitol and glucose media. Moreover, the draft genome sequence with a focus on cellulose related genes was produced. A pH reduction and gluconic acid formation was observed in glucose medium which allowed to produce 6.14 ± 0.02 g/L of bacterial cellulose; the highest bacterial cellulose production obtained was in 1.5% (w/v) mannitol medium (8.77 ± 0.04 g/L), while xylitol provided the lowest (1.35 ± 0.05 g/L) yield. Genomic analysis of K2G30 revealed a peculiar gene sets of cellulose synthase; three bcs operons and a fourth copy of bcsAB gene, that encodes the catalytic core of cellulose synthase. These features can explain the high amount of bacterial cellulose produced by K2G30 strain. Results of this study provide valuable information to industrially exploit acetic acid bacteria in producing bacterial cellulose from different carbon sources including vegetable waste feedstocks containing mannitol.
Keywords: Komagataeibacter xylinus; bacterial cellulose; genome sequencing; gluconic acid; glucose; mannitol; xylitol.
Figures


Similar articles
-
Better under stress: Improving bacterial cellulose production by Komagataeibacter xylinus K2G30 (UMCC 2756) using adaptive laboratory evolution.Front Microbiol. 2022 Oct 12;13:994097. doi: 10.3389/fmicb.2022.994097. eCollection 2022. Front Microbiol. 2022. PMID: 36312960 Free PMC article.
-
Genome sequencing and phylogenetic analysis of K1G4: a new Komagataeibacter strain producing bacterial cellulose from different carbon sources.Biotechnol Lett. 2020 May;42(5):807-818. doi: 10.1007/s10529-020-02811-6. Epub 2020 Jan 25. Biotechnol Lett. 2020. PMID: 31983038
-
How carbon sources drive cellulose synthesis in two Komagataeibacter xylinus strains.Sci Rep. 2024 Sep 3;14(1):20494. doi: 10.1038/s41598-024-71648-0. Sci Rep. 2024. PMID: 39227724 Free PMC article.
-
Biotechnological production of cellulose by acetic acid bacteria: current state and perspectives.Appl Microbiol Biotechnol. 2018 Aug;102(16):6885-6898. doi: 10.1007/s00253-018-9164-5. Epub 2018 Jun 20. Appl Microbiol Biotechnol. 2018. PMID: 29926141 Review.
-
Production of bacterial cellulose from glycerol: the current state and perspectives.Bioresour Bioprocess. 2021 Nov 29;8(1):116. doi: 10.1186/s40643-021-00468-1. Bioresour Bioprocess. 2021. PMID: 38650300 Free PMC article. Review.
Cited by
-
Assessing effectiveness of Komagataeibacter strains for producing surface-microstructured cellulose via guided assembly-based biolithography.Sci Rep. 2021 Sep 29;11(1):19311. doi: 10.1038/s41598-021-98705-2. Sci Rep. 2021. PMID: 34588564 Free PMC article.
-
Study on obtaining bacterial cellulose by Komagataeibacter xylinus in co-culture with lactic acid bacteria in whey.Appl Microbiol Biotechnol. 2025 Aug 21;109(1):191. doi: 10.1007/s00253-025-13582-3. Appl Microbiol Biotechnol. 2025. PMID: 40839214 Free PMC article.
-
Set-Up of Bacterial Cellulose Production From the Genus Komagataeibacter and Its Use in a Gluten-Free Bakery Product as a Case Study.Front Microbiol. 2019 Sep 6;10:1953. doi: 10.3389/fmicb.2019.01953. eCollection 2019. Front Microbiol. 2019. PMID: 31551945 Free PMC article.
-
Bacterial nanocellulose from agro-industrial wastes: low-cost and enhanced production by Komagataeibacter saccharivorans MD1.Sci Rep. 2020 Feb 26;10(1):3491. doi: 10.1038/s41598-020-60315-9. Sci Rep. 2020. PMID: 32103077 Free PMC article.
-
Consecutive bacterial cellulose production by luffa sponge enmeshed with cellulose microfibrils of Acetobacter xylinum under continuous aeration.3 Biotech. 2021 Jan;11(1):6. doi: 10.1007/s13205-020-02569-8. Epub 2021 Jan 2. 3 Biotech. 2021. PMID: 33442505 Free PMC article.
References
-
- Brown R. M. (1996). The biosynthesis of cellulose. J. Macromol. Sci. A 33 1345–1373. 10.1080/10601329608014912 - DOI
LinkOut - more resources
Full Text Sources

