Repurposing of Drugs as Novel Influenza Inhibitors From Clinical Gene Expression Infection Signatures
- PMID: 30761132
- PMCID: PMC6361841
- DOI: 10.3389/fimmu.2019.00060
Repurposing of Drugs as Novel Influenza Inhibitors From Clinical Gene Expression Infection Signatures
Abstract
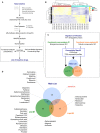
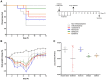
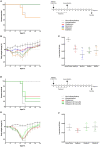
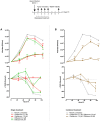
Influenza virus infections remain a major and recurrent public health burden. The intrinsic ever-evolving nature of this virus, the suboptimal efficacy of current influenza inactivated vaccines, as well as the emergence of resistance against a limited antiviral arsenal, highlight the critical need for novel therapeutic approaches. In this context, the aim of this study was to develop and validate an innovative strategy for drug repurposing as host-targeted inhibitors of influenza viruses and the rapid evaluation of the most promising candidates in Phase II clinical trials. We exploited in vivo global transcriptomic signatures of infection directly obtained from a patient cohort to determine a shortlist of already marketed drugs with newly identified, host-targeted inhibitory properties against influenza virus. The antiviral potential of selected repurposing candidates was further evaluated in vitro, in vivo, and ex vivo. Our strategy allowed the selection of a shortlist of 35 high potential candidates out of a rationalized computational screening of 1,309 FDA-approved bioactive molecules, 31 of which were validated for their significant in vitro antiviral activity. Our in vivo and ex vivo results highlight diltiazem, a calcium channel blocker currently used in the treatment of hypertension, as a promising option for the treatment of influenza infections. Additionally, transcriptomic signature analysis further revealed the so far undescribed capacity of diltiazem to modulate the expression of specific genes related to the host antiviral response and cholesterol metabolism. Finally, combination treatment with diltiazem and virus-targeted oseltamivir neuraminidase inhibitor further increased antiviral efficacy, prompting rapid authorization for the initiation of a Phase II clinical trial. This original, host-targeted, drug repurposing strategy constitutes an effective and highly reactive process for the rapid identification of novel anti-infectious drugs, with potential major implications for the management of antimicrobial resistance and the rapid response to future epidemic or pandemic (re)emerging diseases for which we are still disarmed.
Keywords: antivirals; drug repurposing; host targeting; influenza viruses; inhibitors of viral infection; transcriptome.
Figures


Similar articles
-
Drug Repurposing Approaches for the Treatment of Influenza Viral Infection: Reviving Old Drugs to Fight Against a Long-Lived Enemy.Front Immunol. 2019 Mar 19;10:531. doi: 10.3389/fimmu.2019.00531. eCollection 2019. Front Immunol. 2019. PMID: 30941148 Free PMC article. Review.
-
Host-targeted repurposed diltiazem enhances the antiviral activity of direct acting antivirals against Influenza A virus and SARS-CoV-2.Antiviral Res. 2025 May;237:106138. doi: 10.1016/j.antiviral.2025.106138. Epub 2025 Mar 4. Antiviral Res. 2025. PMID: 40049293
-
Inhibition of influenza virus replication by plant-derived isoquercetin.Antiviral Res. 2010 Nov;88(2):227-35. doi: 10.1016/j.antiviral.2010.08.016. Epub 2010 Sep 6. Antiviral Res. 2010. PMID: 20826184
-
Discovery of a Novel Specific Inhibitor Targeting Influenza A Virus Nucleoprotein with Pleiotropic Inhibitory Effects on Various Steps of the Viral Life Cycle.J Virol. 2021 Apr 12;95(9):e01432-20. doi: 10.1128/JVI.01432-20. Print 2021 Apr 12. J Virol. 2021. PMID: 33627391 Free PMC article.
-
Targeting the host or the virus: current and novel concepts for antiviral approaches against influenza virus infection.Antiviral Res. 2012 Dec;96(3):391-404. doi: 10.1016/j.antiviral.2012.09.013. Epub 2012 Sep 26. Antiviral Res. 2012. PMID: 23022351 Free PMC article. Review.
Cited by
-
Response Modifiers: Tweaking the Immune Response Against Influenza A Virus.Front Immunol. 2019 Apr 12;10:809. doi: 10.3389/fimmu.2019.00809. eCollection 2019. Front Immunol. 2019. PMID: 31031778 Free PMC article. Review.
-
Drug Repurposing Approaches for the Treatment of Influenza Viral Infection: Reviving Old Drugs to Fight Against a Long-Lived Enemy.Front Immunol. 2019 Mar 19;10:531. doi: 10.3389/fimmu.2019.00531. eCollection 2019. Front Immunol. 2019. PMID: 30941148 Free PMC article. Review.
-
Antiviral strategies against influenza virus: an update on approved and innovative therapeutic approaches.Cell Mol Life Sci. 2025 Feb 13;82(1):75. doi: 10.1007/s00018-025-05611-1. Cell Mol Life Sci. 2025. PMID: 39945883 Free PMC article. Review.
-
Drug repurposing against SARS-CoV-1, SARS-CoV-2 and MERS-CoV.Future Microbiol. 2021 Nov;16:1341-1370. doi: 10.2217/fmb-2021-0019. Epub 2021 Nov 10. Future Microbiol. 2021. PMID: 34755538 Free PMC article.
-
An Integrated Analysis Reveals Geniposide Extracted From Gardenia jasminoides J.Ellis Regulates Calcium Signaling Pathway Essential for Influenza A Virus Replication.Front Pharmacol. 2021 Nov 19;12:755796. doi: 10.3389/fphar.2021.755796. eCollection 2021. Front Pharmacol. 2021. PMID: 34867371 Free PMC article.
References
-
- Influenza Facts Sheet n°211 (2014) Available online at: http://www.who.int/mediacentre/factsheets/fs211/en/ (Accessed January 20, 2016).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

