e-Sweet: A Machine-Learning Based Platform for the Prediction of Sweetener and Its Relative Sweetness
- PMID: 30761295
- PMCID: PMC6363693
- DOI: 10.3389/fchem.2019.00035
e-Sweet: A Machine-Learning Based Platform for the Prediction of Sweetener and Its Relative Sweetness
Abstract
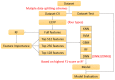
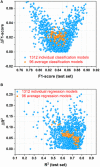
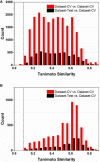
Artificial sweeteners (AS) can elicit the strong sweet sensation with the low or zero calorie, and are widely used to replace the nutritive sugar in the food and beverage industry. However, the safety issue of current AS is still controversial. Thus, it is imperative to develop more safe and potent AS. Due to the costly and laborious experimental-screening of AS, in-silico sweetener/sweetness prediction could provide a good avenue to identify the potential sweetener candidates before experiment. In this work, we curate the largest dataset of 530 sweeteners and 850 non-sweeteners, and collect the second largest dataset of 352 sweeteners with the relative sweetness (RS) from the literature. In light of these experimental datasets, we adopt five machine-learning methods and conformational-independent molecular fingerprints to derive the classification and regression models for the prediction of sweetener and its RS, respectively via the consensus strategy. Our best classification model achieves the 95% confidence intervals for the accuracy (0.91 ± 0.01), precision (0.90 ± 0.01), specificity (0.94 ± 0.01), sensitivity (0.86 ± 0.01), F1-score (0.88 ± 0.01), and NER (Non-error Rate: 0.90 ± 0.01) on the test set, which outperforms the model (NER = 0.85) of Rojas et al. in terms of NER, and our best regression model gives the 95% confidence intervals for the R2(test set) and ΔR2 [referring to |R2(test set)- R2(cross-validation)|] of 0.77 ± 0.01 and 0.03 ± 0.01, respectively, which is also better than the other works based on the conformation-independent 2D descriptors (e.g., 2D Dragon) according to R2(test set) and ΔR2. Our models are obtained by averaging over nineteen data-splitting schemes, and fully comply with the guidelines of Organization for Economic Cooperation and Development (OECD), which are not completely followed by the previous relevant works that are all on the basis of only one random data-splitting scheme for the cross-validation set and test set. Finally, we develop a user-friendly platform "e-Sweet" for the automatic prediction of sweetener and its corresponding RS. To our best knowledge, it is a first and free platform that can enable the experimental food scientists to exploit the current machine-learning methods to boost the discovery of more AS with the low or zero calorie content.
Keywords: QSAR; machine learning method; relative sweetness prediction; sweet taste; sweetener prediction.
Figures
References
LinkOut - more resources
Full Text Sources
Research Materials

