Nuclear pore protein TPR associates with lamin B1 and affects nuclear lamina organization and nuclear pore distribution
- PMID: 30762072
- PMCID: PMC11105453
- DOI: 10.1007/s00018-019-03037-0
Nuclear pore protein TPR associates with lamin B1 and affects nuclear lamina organization and nuclear pore distribution
Abstract
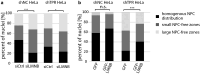
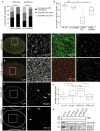
The organization of the nuclear periphery is crucial for many nuclear functions. Nuclear lamins form dense network at the nuclear periphery and play a substantial role in chromatin organization, transcription regulation and in organization of nuclear pore complexes (NPCs). Here, we show that TPR, the protein located preferentially within the nuclear baskets of NPCs, associates with lamin B1. The depletion of TPR affects the organization of lamin B1 but not lamin A/C within the nuclear lamina as shown by stimulated emission depletion microscopy. Finally, reduction of TPR affects the distribution of NPCs within the nuclear envelope and the effect can be reversed by simultaneous knock-down of lamin A/C or the overexpression of lamin B1. Our work suggests a novel role for the TPR at the nuclear periphery: the TPR contributes to the organization of the nuclear lamina and in cooperation with lamins guards the interphase assembly of nuclear pore complexes.
Keywords: Image analysis; Lamina; Lamins; Nuclear pore complex; Nucleus; Super-resolution imaging; TPR; Translocated promoter region.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures

Similar articles
-
Concentric organization of A- and B-type lamins predicts their distinct roles in the spatial organization and stability of the nuclear lamina.Proc Natl Acad Sci U S A. 2019 Mar 5;116(10):4307-4315. doi: 10.1073/pnas.1810070116. Epub 2019 Feb 14. Proc Natl Acad Sci U S A. 2019. PMID: 30765529 Free PMC article.
-
Nuclear envelope remodelling during human spermiogenesis involves somatic B-type lamins and a spermatid-specific B3 lamin isoform.Mol Hum Reprod. 2015 Mar;21(3):225-36. doi: 10.1093/molehr/gau111. Epub 2014 Dec 4. Mol Hum Reprod. 2015. PMID: 25477337
-
Partial cleavage of A-type lamins concurs with their total disintegration from the nuclear lamina during apoptosis.Eur J Cell Biol. 2002 Dec;81(12):677-91. doi: 10.1078/0171-9335-00282. Eur J Cell Biol. 2002. PMID: 12553668
-
Laminopathies: what can humans learn from fruit flies.Cell Mol Biol Lett. 2018 Jul 6;23:32. doi: 10.1186/s11658-018-0093-1. eCollection 2018. Cell Mol Biol Lett. 2018. PMID: 30002683 Free PMC article. Review.
-
Lamins: The backbone of the nucleocytoskeleton interface.Curr Opin Cell Biol. 2024 Feb;86:102313. doi: 10.1016/j.ceb.2023.102313. Epub 2024 Jan 22. Curr Opin Cell Biol. 2024. PMID: 38262116 Review.
Cited by
-
The shifting shape of genomes: dynamics of heterochromatin interactions at the nuclear lamina.Curr Opin Genet Dev. 2021 Apr;67:163-173. doi: 10.1016/j.gde.2021.02.003. Epub 2021 Mar 25. Curr Opin Genet Dev. 2021. PMID: 33774266 Free PMC article. Review.
-
Diamond Nanofilm Normalizes Proliferation and Metabolism in Liver Cancer Cells.Nanotechnol Sci Appl. 2021 Aug 28;14:115-137. doi: 10.2147/NSA.S322766. eCollection 2021. Nanotechnol Sci Appl. 2021. PMID: 34511890 Free PMC article.
-
Protein kinase C activity modulates nuclear Lamin A/C dynamics in HeLa cells.Sci Rep. 2024 Mar 16;14(1):6388. doi: 10.1038/s41598-024-57043-9. Sci Rep. 2024. PMID: 38493209 Free PMC article.
-
Computational analyses reveal spatial relationships between nuclear pore complexes and specific lamins.J Cell Biol. 2021 Apr 5;220(4):e202007082. doi: 10.1083/jcb.202007082. J Cell Biol. 2021. PMID: 33570570 Free PMC article.
-
Nucleoporin TPR Affects C2C12 Myogenic Differentiation via Regulation of Myh4 Expression.Cells. 2021 May 21;10(6):1271. doi: 10.3390/cells10061271. Cells. 2021. PMID: 34063931 Free PMC article.
References
-
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, Leonhardt H, Sedat JW. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320(5881):1332–1336. doi: 10.1126/science.1156947. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

