Alpha protons as NMR probes in deuterated proteins
- PMID: 30762170
- PMCID: PMC6441447
- DOI: 10.1007/s10858-019-00230-y
Alpha protons as NMR probes in deuterated proteins
Abstract
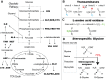
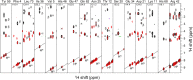
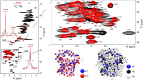
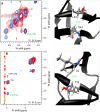
We describe a new labeling method that allows for full protonation at the backbone Hα position, maintaining protein side chains with a high level of deuteration. We refer to the method as alpha proton exchange by transamination (α-PET) since it relies on transaminase activity demonstrated here using Escherichia coli expression. We show that α-PET labeling is particularly useful in improving structural characterization of solid proteins by introduction of an additional proton reporter, while eliminating many strong dipolar couplings. The approach benefits from the high sensitivity associated with 1.3 mm samples, more abundant information including Hα resonances, and the narrow proton linewidths encountered for highly deuterated proteins. The labeling strategy solves amide proton exchange problems commonly encountered for membrane proteins when using perdeuteration and backexchange protocols, allowing access to alpha and all amide protons including those in exchange-protected regions. The incorporation of Hα protons provides new insights, as the close Hα-Hα and Hα-HN contacts present in β-sheets become accessible, improving the chance to determine the protein structure as compared with HN-HN contacts alone. Protonation of the Hα position higher than 90% is achieved for Ile, Leu, Phe, Tyr, Met, Val, Ala, Gln, Asn, Thr, Ser, Glu, Asp even though LAAO is only active at this degree for Ile, Leu, Phe, Tyr, Trp, Met. Additionally, the glycine methylene carbon is labeled preferentially with a single deuteron, allowing stereospecific assignment of glycine alpha protons. In solution, we show that the high deuteration level dramatically reduces R2 relaxation rates, which is beneficial for the study of large proteins and protein dynamics. We demonstrate the method using two model systems, as well as a 32 kDa membrane protein, hVDAC1, showing the applicability of the method to study membrane proteins.
Keywords: Isotopic labeling; L-Amino acid oxidase; Membrane proteins; NMR; Structural restraints; Transamination.
Figures



Similar articles
-
Site-specific protein backbone deuterium 2Hα quadrupolar patterns by proton-detected quadruple-resonance 3D 2HαcαNH MAS NMR spectroscopy.Solid State Nucl Magn Reson. 2023 Jun;125:101861. doi: 10.1016/j.ssnmr.2023.101861. Epub 2023 Mar 21. Solid State Nucl Magn Reson. 2023. PMID: 36989552
-
A strategy to obtain backbone resonance assignments of deuterated proteins in the presence of incomplete amide 2H/1H back-exchange.J Biomol NMR. 2003 Apr;25(4):291-311. doi: 10.1023/a:1023084605308. J Biomol NMR. 2003. PMID: 12766392
-
Hydrogen exchange during cell-free incorporation of deuterated amino acids and an approach to its inhibition.J Biomol NMR. 2011 Dec;51(4):467-76. doi: 10.1007/s10858-011-9575-4. Epub 2011 Oct 8. J Biomol NMR. 2011. PMID: 21984356 Free PMC article.
-
Proton-detected solid-state NMR spectroscopy at aliphatic sites: application to crystalline systems.Acc Chem Res. 2013 Sep 17;46(9):2089-97. doi: 10.1021/ar400063y. Epub 2013 Jun 7. Acc Chem Res. 2013. PMID: 23745638 Review.
-
Proton-Based Ultrafast Magic Angle Spinning Solid-State NMR Spectroscopy.Acc Chem Res. 2017 Apr 18;50(4):1105-1113. doi: 10.1021/acs.accounts.7b00082. Epub 2017 Mar 29. Acc Chem Res. 2017. PMID: 28353338 Free PMC article. Review.
Cited by
-
Proton Detected Solid-State NMR of Membrane Proteins at 28 Tesla (1.2 GHz) and 100 kHz Magic-Angle Spinning.Biomolecules. 2021 May 18;11(5):752. doi: 10.3390/biom11050752. Biomolecules. 2021. PMID: 34069858 Free PMC article.
-
Protein deuteration via algal amino acids to circumvent proton back-exchange for 1H-detected solid-state NMR.Chem Commun (Camb). 2024 Mar 12;60(22):3083-3086. doi: 10.1039/d4cc00213j. Chem Commun (Camb). 2024. PMID: 38407363 Free PMC article.
-
Deuteration of proteins boosted by cell lysates: high-resolution amide and Hα magic-angle-spinning (MAS) NMR without the reprotonation bottleneck.Magn Reson (Gott). 2024 Apr 19;5(1):33-49. doi: 10.5194/mr-5-33-2024. eCollection 2024. Magn Reson (Gott). 2024. PMID: 40384771 Free PMC article.
-
Structure and Gating Behavior of the Human Integral Membrane Protein VDAC1 in a Lipid Bilayer.J Am Chem Soc. 2022 Feb 23;144(7):2953-2967. doi: 10.1021/jacs.1c09848. Epub 2022 Feb 14. J Am Chem Soc. 2022. PMID: 35164499 Free PMC article.
-
Biomolecular solid-state NMR spectroscopy at 1200 MHz: the gain in resolution.J Biomol NMR. 2021 Jul;75(6-7):255-272. doi: 10.1007/s10858-021-00373-x. Epub 2021 Jun 25. J Biomol NMR. 2021. PMID: 34170475 Free PMC article.
References
-
- Agarwal V, Reif B. Residual methyl protonation in perdeuterated proteins for multi-dimensional correlation experiments in MAS solid-state NMR spectroscopy. J Magn Reson. 2008;194:16–24. - PubMed
-
- Agarwal V, Linser R, Fink U, Faelber K, Reif B. Identification of hydroxyl protons, determination of their exchange dynamics, and characterization of hydrogen bonding in a microcrystallin protein. J Am Chem Soc. 2010;132:3187–3195. - PubMed
-
- Akbey Ü, Lange S, Franks WT, Linser R, Rehbein K, Diehl A, Van Rossum BJ, Reif B, Oschkinat H. Optimum levels of exchangeable protons in perdeuterated proteins for proton detection in MAS solid-state NMR spectroscopy. J Biomol NMR. 2010;46:67–73. - PubMed
-
- Andreas LB, Le T, Jaudzems K, Pintacuda G. High-resolution proton-detected NMR of proteins at very fast MAS. J Magn Reson. 2015;253:36–49. - PubMed
-
- Andreas LB, Jaudzems K, Stanek J, Lalli D, Bertarello A, Le Marchand T, Cala-De Paepe D, Kotelovica S, Akopjana I, Knott B, Wegner S, Engelke F, Lesage A, Emsley L, Tars K, Herrmann T, Pintacuda G. Structure of fully protonated proteins by proton-detected magic-angle spinning NMR. Proc. Natl. Acad. Sci. 2016;113:9187–9192. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

