Negative reciprocity, not ordered assembly, underlies the interaction of Sox2 and Oct4 on DNA
- PMID: 30762521
- PMCID: PMC6375704
- DOI: 10.7554/eLife.41017
Negative reciprocity, not ordered assembly, underlies the interaction of Sox2 and Oct4 on DNA
Abstract
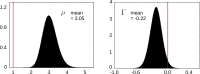
The mode of interaction of transcription factors (TFs) on eukaryotic genomes remains a matter of debate. Single-molecule data in living cells for the TFs Sox2 and Oct4 were previously interpreted as evidence of ordered assembly on DNA. However, the quantity that was calculated does not determine binding order but, rather, energy expenditure away from thermodynamic equilibrium. Here, we undertake a rigorous biophysical analysis which leads to the concept of reciprocity. The single-molecule data imply that Sox2 and Oct4 exhibit negative reciprocity, with expression of Sox2 increasing Oct4's genomic binding but expression of Oct4 decreasing Sox2's binding. Models show that negative reciprocity can arise either from energy expenditure or from a mixture of positive and negative cooperativity at distinct genomic loci. Both possibilities imply unexpected complexity in how TFs interact on DNA, for which single-molecule methods provide novel detection capabilities.
Keywords: Oct4 and Sox2; gene regulation; linear framework; non-equilibrium; none; physics of living systems; single-molecule data.
© 2019, Biddle et al.
Conflict of interest statement
JB, MN, JG No competing interests declared
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

