Differential immune profiles distinguish the mutational subtypes of gastrointestinal stromal tumor
- PMID: 30762585
- PMCID: PMC6486334
- DOI: 10.1172/JCI124108
Differential immune profiles distinguish the mutational subtypes of gastrointestinal stromal tumor
Abstract
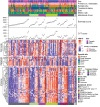
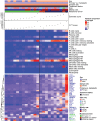
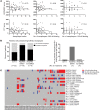
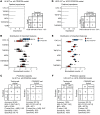
Gastrointestinal stromal tumor (GIST) is the most common human sarcoma, frequently characterized by an oncogenic mutation in the KIT or platelet-derived growth factor receptor alpha (PDGFRA) genes. We performed RNA sequencing of 75 human GIST tumors from 75 patients, comprising the largest cohort of GISTs sequenced to date, in order to discover differences in the immune infiltrates of KIT and PDGFRA-mutant GIST. Through bioinformatics, immunohistochemistry, and flow cytometry, we found that PDGFRA-mutant GISTs harbored more immune cells with increased cytolytic activity when compared to KIT-mutant GISTs. PDGFRA-mutant GISTs expressed many chemokines, such as CXCL14, at a significantly higher level when compared to KIT-mutant GISTs and exhibited more diverse driver-derived neoepitope:HLA binding, both of which may contribute to PDGFRA-mutant GIST immunogenicity. Through machine learning, we generated gene expression-based immune profiles capable of differentiating KIT and PDGFRA-mutant GISTs, and also identified additional immune features of high PD-1 and PD-L1 expressing tumors across all GIST mutational subtypes, which may provide insight into immunotherapeutic opportunities and limitations in GIST.
Keywords: Bioinformatics; Cancer immunotherapy; Immunology; Oncogenes; Oncology.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

