TyrR is involved in the transcriptional regulation of biofilm formation and D-alanine catabolism in Azospirillum brasilense Sp7
- PMID: 30763337
- PMCID: PMC6375630
- DOI: 10.1371/journal.pone.0211904
TyrR is involved in the transcriptional regulation of biofilm formation and D-alanine catabolism in Azospirillum brasilense Sp7
Abstract
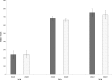
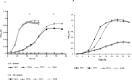
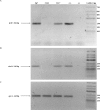
Azospirillum brasilense is one of the most studied species of diverse agronomic plants worldwide. The benefits conferred to plants inoculated with Azospirillum have been primarily attributed to its capacity to fix atmospheric nitrogen and synthesize phytohormones, especially indole-3-acetic acid (IAA). The principal pathway for IAA synthesis involves the intermediate metabolite indole pyruvic acid. Successful colonization of plants by Azospirillum species is fundamental to the ability of these bacteria to promote the beneficial effects observed in plants. Biofilm formation is an essential step in this process and involves interactions with the host plant. In this study, the tyrR gene was cloned, and the translated product was observed to exhibit homology to TyrR protein, a NtrC/NifA-type activator. Structural studies of TyrR identified three putative domains, including a domain containing binding sites for aromatic amino acids in the N-terminus, a central AAA+ ATPase domain, and a helix-turn-helix DNA binding motif domain in the C-terminus, which binds DNA sequences in promoter-operator regions. In addition, a bioinformatic analysis of promoter sequences in A. brasilense Sp7 genome revealed that putative promoters encompass one to three TyrR boxes in genes predicted to be regulated by TyrR. To gain insight into the phenotypes regulated by TyrR, a tyrR-deficient strain derived from A. brasilense Sp7, named A. brasilense 2116 and a complemented 2116 strain harboring a plasmid carrying the tyrR gene were constructed. The observed phenotypes indicated that the putative transcriptional regulator TyrR is involved in biofilm production and is responsible for regulating the utilization of D-alanine as carbon source. In addition, TyrR was observed to be absolutely required for transcriptional regulation of the gene dadA encoding a D-amino acid dehydrogenase. The data suggested that TyrR may play a major role in the regulation of genes encoding a glucosyl transferase, essential signaling proteins, and amino acids transporters.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Argaet VP, Wilson TJ, Davidson BE. Purification of the Escherichia coli regulatory protein TyrR and analysis of its interactions with ATP, tyrosine, phenylalanine, and tryptophan. The Journal of biological chemistry. 1994;269(7):5171–8. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

