Comparative analysis of mutational robustness of the intrinsically disordered viral protein VPg and of its interactor eIF4E
- PMID: 30763345
- PMCID: PMC6375565
- DOI: 10.1371/journal.pone.0211725
Comparative analysis of mutational robustness of the intrinsically disordered viral protein VPg and of its interactor eIF4E
Abstract
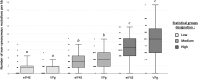
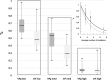
Conformational intrinsic disorder is a feature present in many virus proteins. Intrinsically disordered regions (IDRs) have weaker structural requirement than ordered regions and mutations in IDRs could have a lower impact on the virus fitness. This could favor its exploration of adaptive solutions. The potyviral protein VPg contains IDRs with determinants for adaptation to its host plant. To experimentally assess whether IDRs are more resistant to mutations than ordered regions, the biologically relevant interaction between mutant libraries of both VPg and the eukaryotic translation initiation factor 4E (eIF4E) and their respective wild type partner was examined using yeast two hybrid assay. Our data shows that VPg is significantly more robust to mutations than eIF4E and as such belongs to a particular class of intrinsically disordered proteins. This result is discussed from the standpoint of IDRs involvement in the virus adaptive processes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg.Plant J. 2008 Apr;54(1):56-68. doi: 10.1111/j.1365-313X.2008.03407.x. Epub 2008 Jan 7. Plant J. 2008. PMID: 18182024
-
Hydrodynamic Behavior of the Intrinsically Disordered Potyvirus Protein VPg, of the Translation Initiation Factor eIF4E and of their Binary Complex.Int J Mol Sci. 2019 Apr 11;20(7):1794. doi: 10.3390/ijms20071794. Int J Mol Sci. 2019. PMID: 30978975 Free PMC article.
-
Helper component proteinase of the genus Potyvirus is an interaction partner of translation initiation factors eIF(iso)4E and eIF4E and contains a 4E binding motif.J Virol. 2011 Jul;85(13):6784-94. doi: 10.1128/JVI.00485-11. Epub 2011 Apr 27. J Virol. 2011. PMID: 21525344 Free PMC article.
-
The Molecular Maze of Potyviral and Host Protein Interactions.Annu Rev Virol. 2024 Sep;11(1):147-170. doi: 10.1146/annurev-virology-100422-034124. Epub 2024 Aug 30. Annu Rev Virol. 2024. PMID: 38848589 Review.
-
The Disorderly Nature of Caliciviruses.Viruses. 2024 Aug 19;16(8):1324. doi: 10.3390/v16081324. Viruses. 2024. PMID: 39205298 Free PMC article. Review.
Cited by
-
Insights into the Functions of eIF4E-Biding Motif of VPg in Potato Virus A Infection.Viruses. 2020 Feb 11;12(2):197. doi: 10.3390/v12020197. Viruses. 2020. PMID: 32053987 Free PMC article.
-
Evolutionary Forces and Codon Bias in Different Flavors of Intrinsic Disorder in the Human Proteome.J Mol Evol. 2020 Mar;88(2):164-178. doi: 10.1007/s00239-019-09921-4. Epub 2019 Dec 10. J Mol Evol. 2020. PMID: 31820049
-
New technologies to analyse protein function: an intrinsic disorder perspective.F1000Res. 2020 Feb 10;9:F1000 Faculty Rev-101. doi: 10.12688/f1000research.20867.1. eCollection 2020. F1000Res. 2020. PMID: 32089835 Free PMC article. Review.
-
Deciphering Cowpea Resistance to Potyvirus: Assessment of eIF4E Gene Mutations and Their Impact on the eIF4E-VPg Protein Interaction.Viruses. 2025 Jul 28;17(8):1050. doi: 10.3390/v17081050. Viruses. 2025. PMID: 40872765 Free PMC article.
-
Evolution of intrinsic disorder in the structural domains of viral and cellular proteomes.Sci Rep. 2025 Jan 22;15(1):2878. doi: 10.1038/s41598-025-86045-4. Sci Rep. 2025. PMID: 39843714 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

