(Ph)ighting Phages: How Bacteria Resist Their Parasites
- PMID: 30763533
- PMCID: PMC6383810
- DOI: 10.1016/j.chom.2019.01.009
(Ph)ighting Phages: How Bacteria Resist Their Parasites
Abstract
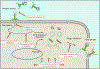
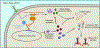
Bacteria are under constant attack from bacteriophages (phages), bacterial parasites that are the most abundant biological entity on earth. To resist phage infection, bacteria have evolved an impressive arsenal of anti-phage systems. Recent advances have significantly broadened and deepened our understanding of how bacteria battle phages, spearheaded by new systems like CRISPR-Cas. This review aims to summarize bacterial anti-phage mechanisms, with an emphasis on the most recent developments in the field.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures


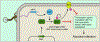
References
-
- Alawneh AM, Qi D, Yonesaki T, and Otsuka Y (2016). An ADP-ribosyltransferase Alt of bacteriophage T4 negatively regulates the Escherichia coli MazF toxin of a toxin-antitoxin module. Mol Microbiol 99, 188–198. - PubMed
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, and Horvath P (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. - PubMed
-
- Bingham R, Ekunwe SI, Falk S, Snyder L, and Kleanthous C (2000). The major head protein of bacteriophage T4 binds specifically to elongation factor Tu. J Biol Chem 275, 23219–23226. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

