Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis
- PMID: 30763538
- PMCID: PMC6885004
- DOI: 10.1016/j.chom.2019.01.008
Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis
Abstract
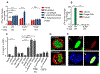
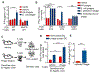
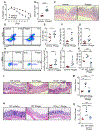
Bacteriophages are the most abundant members of the microbiota and have the potential to shape gut bacterial communities. Changes to bacteriophage composition are associated with disease, but how phages impact mammalian health remains unclear. We noted an induction of host immunity when experimentally treating bacterially driven cancer, leading us to test whether bacteriophages alter immune responses. Treating germ-free mice with bacteriophages leads to immune cell expansion in the gut. Lactobacillus, Escherichia, and Bacteroides bacteriophages and phage DNA stimulated IFN-γ via the nucleotide-sensing receptor TLR9. The resultant immune responses were both phage and bacteria specific. Additionally, increasing bacteriophage levels exacerbated colitis via TLR9 and IFN-γ. Similarly, ulcerative colitis (UC) patients responsive to fecal microbiota transplantation (FMT) have reduced phages compared to non-responders, and mucosal IFN-γ positively correlates with bacteriophage levels. Bacteriophages from active UC patients induced more IFN-γ compared to healthy individuals. Collectively, these results indicate that bacteriophages can alter mucosal immunity to impact mammalian health.
Keywords: FMT; IFN-y; T cells; TLR9; Th1; bacteriophages; colitis; dendritic cells.
Published by Elsevier Inc.
Conflict of interest statement
DECLARATION OF INTEREST
The authors declare no conflict of interest.
Figures




Comment in
-
The Innate Sense of Bacteriophages.Cell Host Microbe. 2019 Feb 13;25(2):177-179. doi: 10.1016/j.chom.2019.01.020. Cell Host Microbe. 2019. PMID: 30763530
-
Gut phages at the centre.Nat Rev Microbiol. 2019 Apr;17(4):195. doi: 10.1038/s41579-019-0174-9. Nat Rev Microbiol. 2019. PMID: 30820034 No abstract available.
References
-
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, and Clevers H (2009). Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611. - PubMed
-
- Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, Stotland A, Wolkowicz R, Cutting AS, Doran KS, et al. (2013). Bacteriophage adhering to mucus provide a nonhost-derived immunity. Proceedings of the National Academy of Sciences of the United States of America 110, 10771–10776. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

