Scavenger Receptor Class A1 Mediates Uptake of Morpholino Antisense Oligonucleotide into Dystrophic Skeletal Muscle
- PMID: 30763772
- PMCID: PMC6374502
- DOI: 10.1016/j.omtn.2019.01.008
Scavenger Receptor Class A1 Mediates Uptake of Morpholino Antisense Oligonucleotide into Dystrophic Skeletal Muscle
Abstract
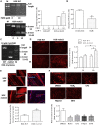
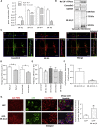
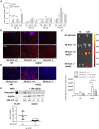
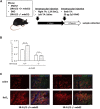
Exon skipping using phosphorodiamidate morpholino oligomers (PMOs) is a promising treatment strategy for Duchenne muscular dystrophy (DMD). The most significant limitation of these clinically used compounds is their lack of delivery systems that target muscles; thus, cell-penetrating peptides are being developed to enhance uptake into muscles. Recently, we reported that uptake of peptide-conjugated PMOs into myofibers was mediated by scavenger receptor class A (SR-A), which binds negatively charged ligands. However, the mechanism by which the naked PMOs are taken up into fibers is poorly understood. In this study, we found that PMO uptake and exon-skipping efficiency were promoted in dystrophin-deficient myotubes via endocytosis through a caveolin-dependent pathway. Interestingly, SR-A1 was upregulated and localized in juxtaposition with caveolin-3 in these myotubes and promoted PMO-induced exon skipping. SR-A1 was also upregulated in the skeletal muscle of mdx52 mice and mediated PMO uptake. In addition, PMOs with neutral backbones had negative zeta potentials owing to their nucleobase compositions and interacted with SR-A1. In conclusion, PMOs with negative zeta potential were taken up into dystrophin-deficient skeletal muscle by upregulated SR-A1. Therefore, the development of a drug delivery system targeting SR-A1 could lead to highly efficient exon-skipping therapies for DMD.
Keywords: Duchenne muscular dystrophy; delivery; endocytosis; exon skipping; mdx52; oligonucleotide uptake; phosphorodiamidate morpholino oligomer; scavenger receptor; splice switching; zeta potential.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Arora V., Devi G.R., Iversen P.L. Neutrally charged phosphorodiamidate morpholino antisense oligomers: uptake, efficacy and pharmacokinetics. Curr. Pharm. Biotechnol. 2004;5:431–439. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

