Epigenetically Altered T Cells Contribute to Lupus Flares
- PMID: 30764520
- PMCID: PMC6406295
- DOI: 10.3390/cells8020127
Epigenetically Altered T Cells Contribute to Lupus Flares
Abstract
Lupus flares when genetically predisposed people encounter exogenous agents such as infections and sun exposure and drugs such as procainamide and hydralazine, but the mechanisms by which these agents trigger the flares has been unclear. Current evidence indicates that procainamide and hydralazine, as well as inflammation caused by the environmental agents, can cause overexpression of genes normally silenced by DNA methylation in CD4⁺ T cells, converting them into autoreactive, proinflammatory cytotoxic cells that are sufficient to cause lupus in mice, and similar cells are found in patients with active lupus. More recent studies demonstrate that these cells comprise a distinct CD4⁺ T cell subset, making it a therapeutic target for the treatment of lupus flares. Transcriptional analyses of this subset reveal proteins uniquely expressed by this subset, which may serve as therapeutic to deplete these cells, treating lupus flares.
Keywords: DNA methylation; environment; epigenetics; lupus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
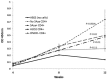
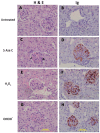
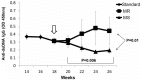
Comment in
-
Commentary on "Epigenetically Altered T Cells Contribute to Lupus Flares".J Cell Immunol. 2019;1(1):10.33696/immunology.1.004. doi: 10.33696/immunology.1.004. J Cell Immunol. 2019. PMID: 31737868 Free PMC article. No abstract available.
Similar articles
-
Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs.J Immunol. 2005 May 15;174(10):6212-9. doi: 10.4049/jimmunol.174.10.6212. J Immunol. 2005. PMID: 15879118
-
The interaction between environmental triggers and epigenetics in autoimmunity.Clin Immunol. 2018 Jul;192:1-5. doi: 10.1016/j.clim.2018.04.005. Epub 2018 Apr 9. Clin Immunol. 2018. PMID: 29649575 Free PMC article. Review.
-
Environmental exposures, epigenetic changes and the risk of lupus.Lupus. 2014 May;23(6):568-76. doi: 10.1177/0961203313499419. Lupus. 2014. PMID: 24763540 Free PMC article. Review.
-
CD4(+) T cells epigenetically modified by oxidative stress cause lupus-like autoimmunity in mice.J Autoimmun. 2015 Aug;62:75-80. doi: 10.1016/j.jaut.2015.06.004. Epub 2015 Jul 9. J Autoimmun. 2015. PMID: 26165613 Free PMC article.
-
Dissecting complex epigenetic alterations in human lupus.Arthritis Res Ther. 2013 Jan 29;15(1):201. doi: 10.1186/ar4125. Arthritis Res Ther. 2013. PMID: 23374884 Free PMC article. Review.
Cited by
-
Epigenetic Contribution and Genomic Imprinting Dlk1-Dio3 miRNAs in Systemic Lupus Erythematosus.Genes (Basel). 2021 May 1;12(5):680. doi: 10.3390/genes12050680. Genes (Basel). 2021. PMID: 34062726 Free PMC article. Review.
-
Mesenchymal Stem Cells Activate the MEK/ERK Signaling Pathway and Enhance DNA Methylation via DNMT1 in PBMC from Systemic Lupus Erythematosus.Biomed Res Int. 2020 Nov 17;2020:4174082. doi: 10.1155/2020/4174082. eCollection 2020. Biomed Res Int. 2020. PMID: 33282947 Free PMC article.
-
Epigenetic Mechanisms and Posttranslational Modifications in Systemic Lupus Erythematosus.Int J Mol Sci. 2019 Nov 13;20(22):5679. doi: 10.3390/ijms20225679. Int J Mol Sci. 2019. PMID: 31766160 Free PMC article. Review.
-
A fair machine learning model to predict flares of systemic lupus erythematosus.JAMIA Open. 2025 Jul 26;8(4):ooaf072. doi: 10.1093/jamiaopen/ooaf072. eCollection 2025 Aug. JAMIA Open. 2025. PMID: 40718760 Free PMC article.
-
Methyl donor micronutrients, CD40-ligand methylation and disease activity in systemic lupus erythematosus: A cross-sectional association study.Lupus. 2021 Oct;30(11):1773-1780. doi: 10.1177/09612033211034559. Epub 2021 Jul 20. Lupus. 2021. PMID: 34284675 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

