The transcription factor Duxbl mediates elimination of pre-T cells that fail β-selection
- PMID: 30765463
- PMCID: PMC6400535
- DOI: 10.1084/jem.20181444
The transcription factor Duxbl mediates elimination of pre-T cells that fail β-selection
Abstract
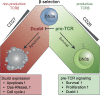
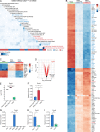
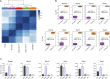
T cell development is critically dependent on successful rearrangement of antigen-receptor chains. At the β-selection checkpoint, only cells with a functional rearrangement continue in development. However, how nonselected T cells proceed in their dead-end fate is not clear. We identified low CD27 expression to mark pre-T cells that have failed to rearrange their β-chain. Expression profiling and single-cell transcriptome clustering identified a developmental trajectory through β-selection and revealed specific expression of the transcription factor Duxbl at a stage of high recombination activity before β-selection. Conditional transgenic expression of Duxbl resulted in a developmental block at the DN3-to-DN4 transition due to reduced proliferation and enhanced apoptosis, whereas RNA silencing of Duxbl led to a decrease in apoptosis. Transcriptome analysis linked Duxbl to elevated expression of the apoptosis-inducing Oas/RNaseL pathway. RNaseL deficiency or sustained Bcl2 expression led to a partial rescue of cells in Duxbl transgenic mice. These findings identify Duxbl as a regulator of β-selection by inducing apoptosis in cells with a nonfunctional rearrangement.
© 2019 Klein et al.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

