Local frustration around enzyme active sites
- PMID: 30765513
- PMCID: PMC6410768
- DOI: 10.1073/pnas.1819859116
Local frustration around enzyme active sites
Abstract
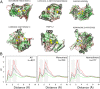
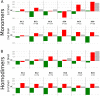
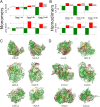
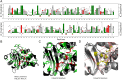
Conflicting biological goals often meet in the specification of protein sequences for structure and function. Overall, strong energetic conflicts are minimized in folded native states according to the principle of minimal frustration, so that a sequence can spontaneously fold, but local violations of this principle open up the possibility to encode the complex energy landscapes that are required for active biological functions. We survey the local energetic frustration patterns of all protein enzymes with known structures and experimentally annotated catalytic residues. In agreement with previous hypotheses, the catalytic sites themselves are often highly frustrated regardless of the protein oligomeric state, overall topology, and enzymatic class. At the same time a secondary shell of more weakly frustrated interactions surrounds the catalytic site itself. We evaluate the conservation of these energetic signatures in various family members of major enzyme classes, showing that local frustration is evolutionarily more conserved than the primary structure itself.
Keywords: bioinformatics; catalytic sites; evolution; local frustration; protein enzymes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Bar-Even A, Milo R, Noor E, Tawfik DS. The moderately efficient enzyme: Futile encounters and enzyme floppiness. Biochemistry. 2015;54:4969–4977. - PubMed
-
- Meiering EM, Serrano L, Fersht AR. Effect of active site residues in barnase on activity and stability. J Mol Biol. 1992;225:585–589. - PubMed
-
- Sanchez I, Tejero J, Gomez-Moreno C, Medina M, Serrano L. Point mutations in protein globular domains: Contributions from function, stability and misfolding. J Mol Biol. 2006;363:422–432. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

